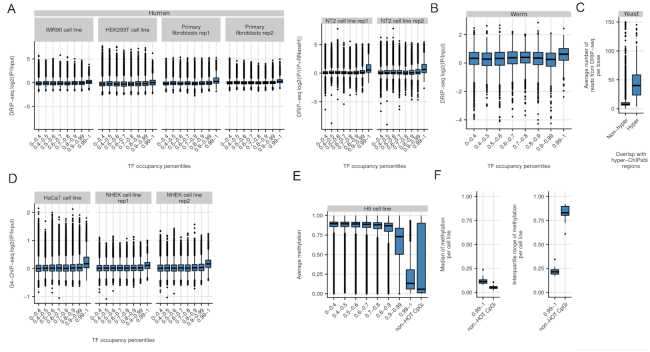
Figure 4.
R-loops are associated with HOT regions. The boxplots show DRIP-seq log2(IP/control) for HOT regions and control regions binned based on their TF occupancy percentile in (A) Various human cell lines and in (B) worm. Boxplots show DRIP-seq read count per base-pair for hyper-ChIPable regions and all other genes as controls. (C) HyperChIP-able regions in yeast are enriched in R-loops. (D) HOT regions are enriched with G-quadruplex DNA (G4-ChIP-seq). Boxplots show log2(IP/control) for HOT regions and control regions binned based on their TF occupancy percentile. (E) HOT regions are hypo-methylated in comparison to controls in H9 cell line. Boxplots show distributions of methylation for HOT regions (rightmost boxplot) and control regions binned based on their TF occupancy percentile. (F) Left boxplot shows distributions of methylation medians across cell types for HOT regions and CpG islands that are not associated with HOT regions (non-HOT CpGi). Right boxplot shows distributions of methylation IQRs (interquartile ranges) across cell types for HOT regions and non-HOT CpGi.