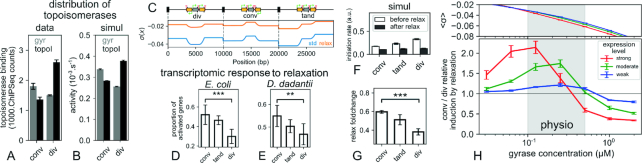
Figure 4.
(A) ChIP-Seq data show that topoisomerase I and the DNA gyrase preferentially bind in divergent and convergent regions, respectively (43). (B) The observed distribution naturally emerges from our modeling of TSC in physiologically relevant conditions (moderate gene expression and 0.25 μM gyrase concentration, see Materials and Methods). (C) Simulations were carried on a model genome with three distinct topological domains, each carrying two active genes (yellow) in different orientations, and a central inactive gene (gray) as a regulatory probe. The heterogeneous SC levels are shown before (blue) and after (orange) relaxation by gyrase inhibition, and they give rise to the heterogeneous recruitment of topoisomerases observed in B. (D) Transcriptomics data in Escherichia coli from (9) show that genes’ response to chromosomal relaxation is tightly related to their local orientation. (E) The same is observed in new RNA-Seq data from Dickeya dadantii, in exponential as well as at transition to stationary (Supplementary Figure S4B) phase, confirming that this feature is not organism- or condition-specific. (F) Simulating the action of the antibiotics exhibits a different effect on the promoters’ initiation rates, depending on their local orientation. (G) As a result, relaxation favours convergent genes versus divergent ones, in agreement with genome-wide experimental data. (H) The average SC level (upper panel) and relative convergent/divergent foldchange due to relaxation (lower panel) was computed for a range of initial DNA gyrase concentrations and expression strengths (of all active genes), corresponding to effective waiting times of 1, 2.5 and 10 min between transcription events at each promoter, respectively. Data shown in B, C, F and G correspond to the central datapoint with moderate expression.