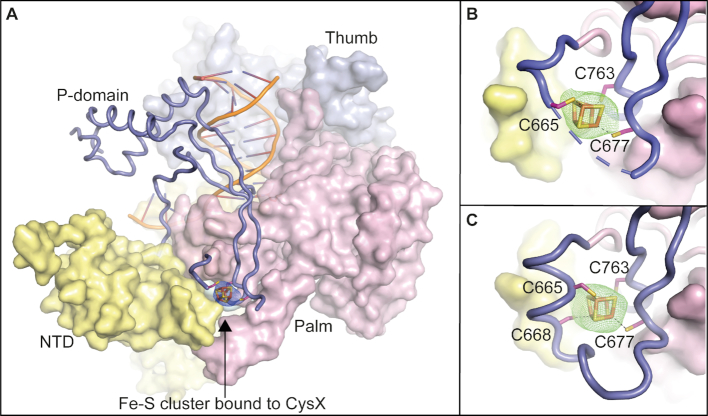
Figure 2.
Crystal structures of the catalytic core of Pol ϵ showing an Fe–S cluster bound to the CysX motif. (A) Overall structure of Pol2CORE (1–1228) with an [4Fe–4S] cluster modelled at the base of the P-domain (PDB ID: 6h1v). An anomalous map over the Fe–S cluster, shown in blue mesh at 3σ supports the presence of a cubic Fe–S cluster. Pol2CORE (1–1228) was over-expressed in yeast. The P-domain is shown in slate blue cartoon. The N-terminal, palm, and thumb domains are in yellow, pink, and light blue surface representations, respectively. (B) Close-up view of the Fe–S cluster bound to the CysX motif at the base of the P-domain (PDB ID: 6h1v). An omit map at 3σ is shown in green mesh. (C) The crystal structure of Pol2CORE (1–1187, PDB ID: 6qib) shows the complete loop region folded at the base of the P-domain revealing the positions of all four cysteines in the CysX motif. Pol2CORE (1–1187) was over-expressed in E. coli.