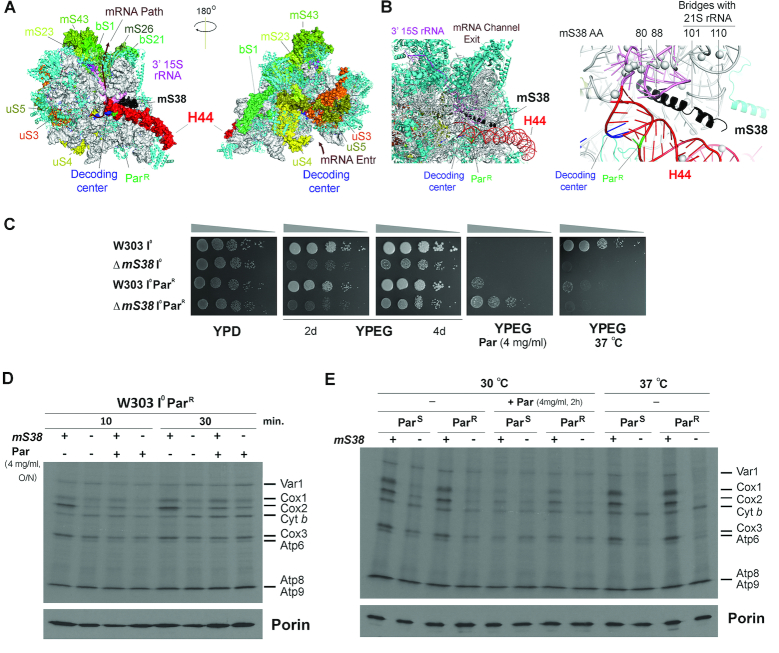
Figure 6.
mS38 genetically interacts with mutations in the 15S rRNA helix 44. See also Supplementary Figure S6. (A) Localization of the mRNA path in the mtSSU, the proteins located at the entrance and exit channels, as well as mS38, the 15S rRNA helix 44 and the decoding center in the yeast mitoribosome structure (PDB 5MRC) (8). (B) Close-up of the mS38-h44 region in the mtSSU structure. The location of the paromomycin-resistance mutation (ParR) and key amino acids in mS38 are indicated. (C) Serial dilutions growth test of the indicated strains in complete fermentable (YPD) or non-fermentable (YPEG) solid media. The effects of paromomycin supplementation and incubation at 37°C were also tested. Pictures were taken after two days of growth or the time indicated. (D, E) In vivo mitochondrial protein synthesis of the indicated strains performed as in Figure 2D. In (D), the intronless WT (W303 I0) and ΔmS38 strains with mtDNA carrying the paromomycin-resistant mutation (ParR) were incubated or not overnight (ON) in the presence of 4 mg/ml of paromomycin (Par) and pulsed with 35S-methionine for 10 or 30 min. In (E), the strains with mtDNA carrying paromomycin-sensitive or resistant 15S rRNA sequences were incubated at 30°C in the presence or absence of 4 mg/ml paromomycin for 2 hrs or 37°C and pulsed for 10 minutes with 35S-methionine in the same conditions.