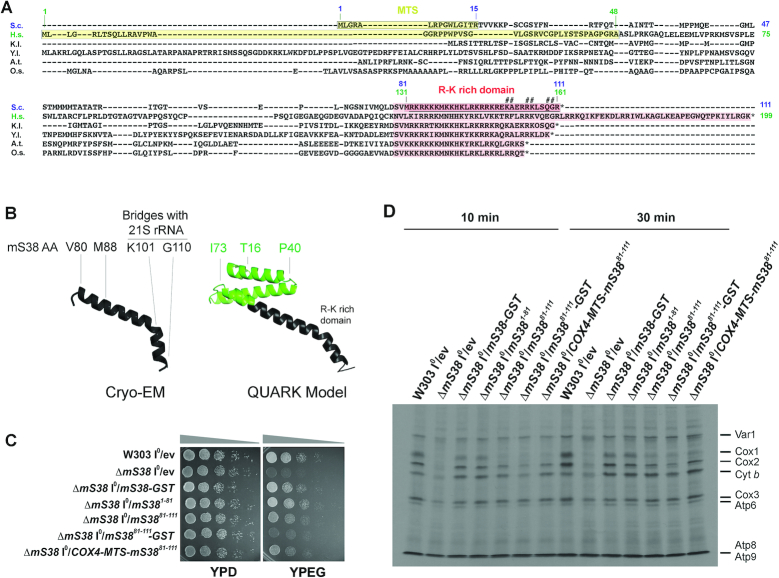
Figure 7.
The N- and C-terminal portions of mS38 perform different roles in ribosome assembly and mRNA-specific translation. See also Supplementary Figures S7 and S8. (A) Alignment of mS38 sequences from Saccharomyces cerevisiae (S.c.), Homo sapiens (H.s.), other yeasts (Kluyveromyces lactis—K.l- and Yarrowia lipolytica—Y.l.) and plants (Arabidopsis thaliana—A.t., Oriza sativa—O.s.). The mitochondrial targeting sequence (MTS) in S.c and H.s. are marked in yellow. The arginine-lysine (R-G)-rich domain present in the C-terminus of all proteins is labeled in red. Numbers in blue indicate amino acids (aa) in the S. cerevisiae protein and numbers in green, amino acids in the human protein. The underlined region in S. cerevisiae mS38 is the region that was resolved in the cryo-EM structural reconstruction by (8). The # symbol indicates residues in S. cerevisiae mS38 involved in the formation of intersubunit bridge mB3 with elements of the 21S rRNA (8). Bridge mB3 is exclusive of mitochondrial ribosomes but conserved from yeast to mammals. (B) Structure of S. cerevisiae mS38 as obtained by cryo-EM including amino acids 80–111 (8) and a model of the whole mature protein (aa 16–111) obtained using Quark, a computer algorithm for ab initio protein structure prediction (39). Key amino acid residues are indicated. (C) Serial dilutions growth test of the indicated strains in complete media containing fermentable (YPD) or non-fermentable (YPEG) carbon sources. Pictures were taken after two days of growth at 30°C. (D) In vivo mitochondrial protein synthesis using the indicated strains and performed as in Figure 2D following 35S-methionine pulses of the indicated increasing times.