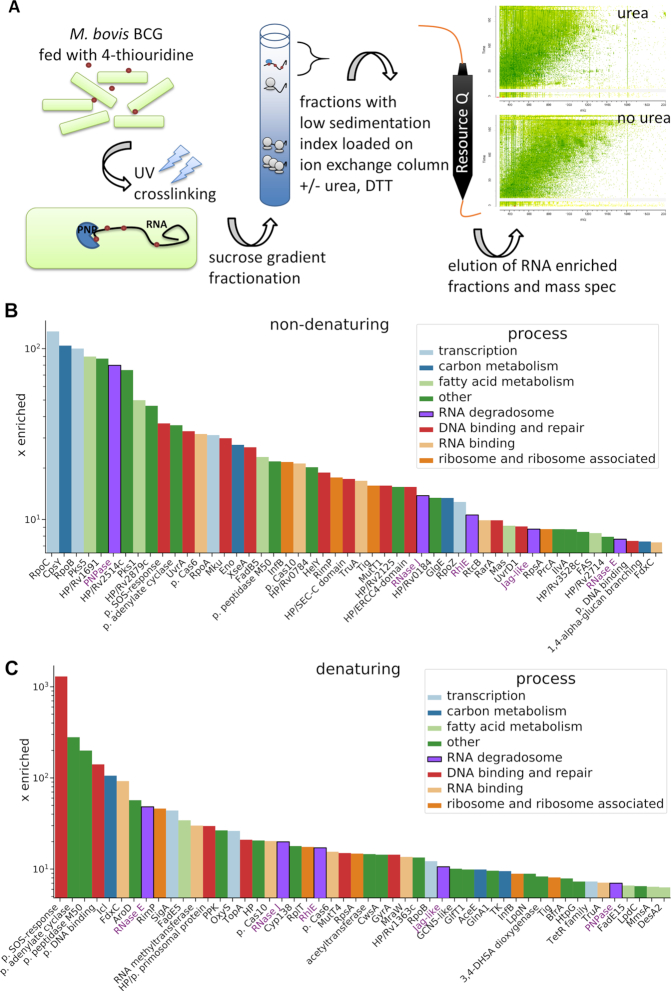
Figure 1.
Analysis of the RNA-bound proteome of M. bovis BCG. (A) Experiment schematic. Labelled RNA is UV-crosslinked with protein, which is then purified using a sucrose gradient followed by anion exchange chromatography. Purified proteins were analysed with mass spectrometry. Fifty candidate RNA binding proteins present in preparations under non-denaturing (B) and denaturing (C) conditions were chosen based on their enrichment factor (x enriched) in comparison to the intracellular levels in M. tuberculosis (Supplementary Table S1, Peptide Atlas, (32)). The MaxQuant intensity values were transformed into ppm values by calculating the relative abundance of each protein comparing to the total abundance of the entire sample. Next, the individual ppm values for each protein in the RNA–protein pull-down experiments were divided by corresponding ppm values for intracellular abundance. The putative RNA degradosome constituents enriched under both conditions are annotated with purple font.