Figure 4.
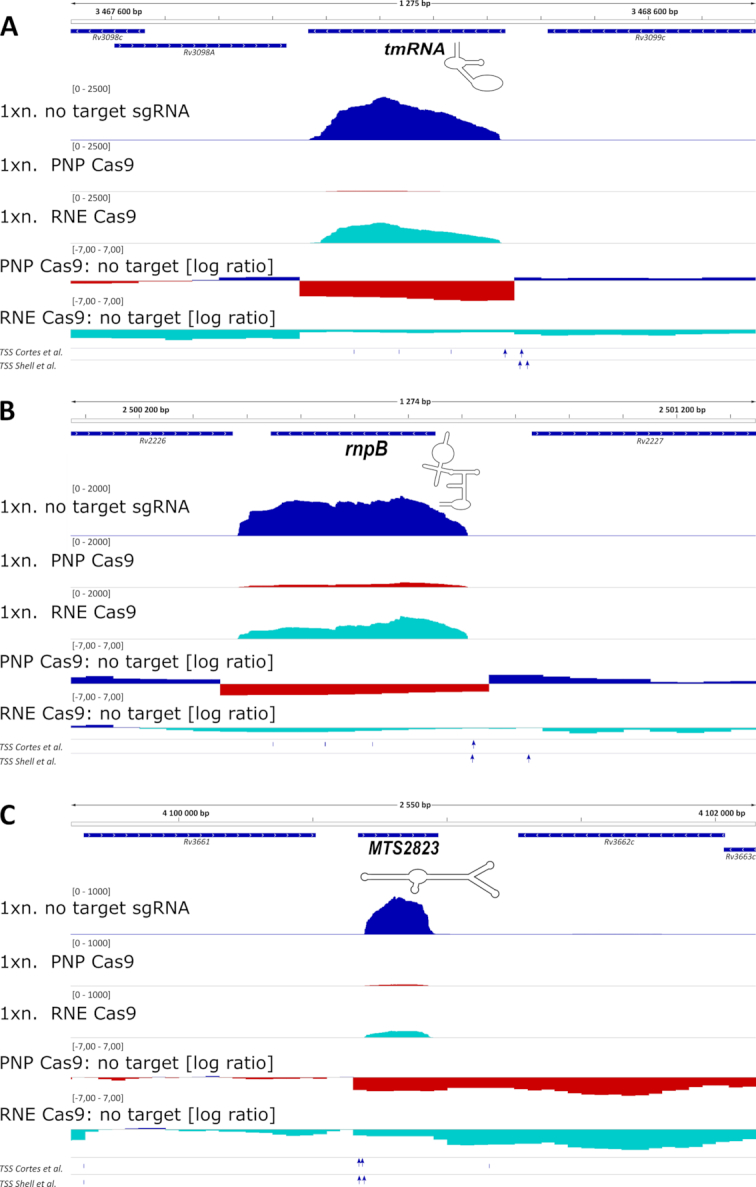
Expression of important non-coding RNAs is affected by the removal of PNPase and RNase E. Expression profiles of important non-coding RNAs that were changed significantly in the total RNA Seq results for PNP Cas9 strain: (A) transfer messenger RNA - tmRNA, (B) RNA component of RNase P – rnpB and (C) MTS2823, mycobacterial 6S-like molecule are shown. The transcripts were depleted to a lesser level from the RNE Cas9 transcriptomes, with only MTS2823 passing the cut-off values set on false discovery rate (0.05) and log2 fold change (|1.585|). 1× normalized expression profiles (1xn.; as bedGraph tracks) are presented together with log2 expression ratios for the chosen genomic regions. Gene annotations and experimentally verified transcription start sites (TSS) are included underneath each genomic segment. TSSs specific to mature ncRNAs are represented by blue arrows.
