Fig. 1. Pleiotropy is central to our understanding of mammalian gene function.
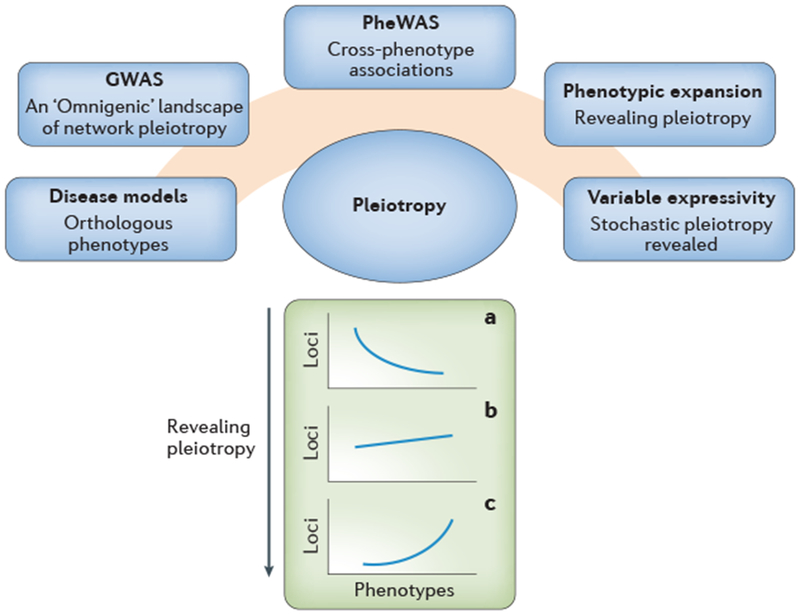
Pleiotropy, the multiple functions of a gene, is manifest through the exploration of disease models and a variety of other phenomena. These include genome-wide association studies (GWAS) where for complex traits the association signals are widely spread across numerous genes and not simply in core disease pathways. The implication is that network pleiotropy is rife and that all genes expressed in a particular tissue are likely to affect phenotype outcome – the “omingenic” hypothesis16. Pleiotropy is also revealed in phenome-wide association studies (PheWAS) where the associations of individual genetic variants with multiple phenotypes, known as cross-phenotype associations, are uncovered17. The well-known phenomenon of phenotypic expansion in human genetics also exemplifies the pervasiveness of human pleiotropy15. Finally, the well-known phenomenon of variable expressivity by which the expression of different aspects of phenotype varies across individuals with identical genotype is also revealing of pleiotropy. Uncovering pleiotropy to its fullest extent is a critical ambition for high-throughput mouse phenomics with the aim of improving our knowledge of pleiotropy and developing datasets where multiple functions are documented. Currently, for most loci we have limited knowledge of pleiotropy and for most genes our knowledge of phenotypes is limited (a). The challenge for genetics is to extend our knowledge of the multiple functions of genes to an increasing number of loci (b) and ultimately to most of the genes in the genome (c).
