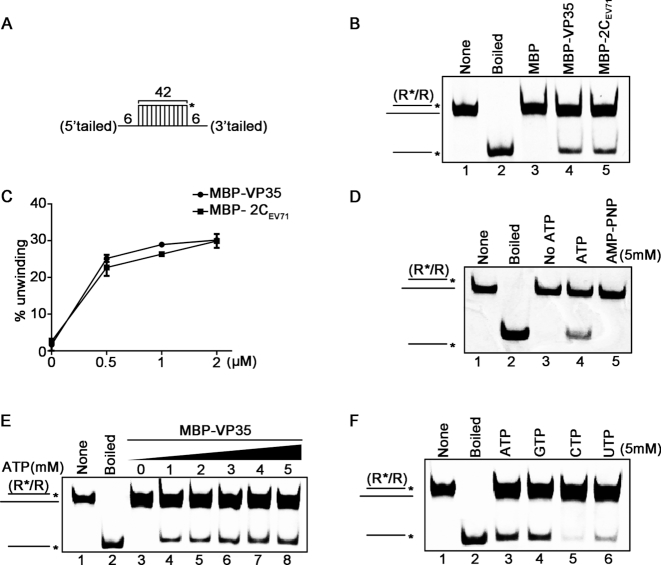
Figure 2.
EBOV VP35 has the NTP-dependent RNA helix unwinding activity. (A) Schematic illustration of the standard RNA helix substrate (R*/R). Asterisks indicate the HEX-labeled strands. (B) The standard RNA helix substrate (0.1 pmol) was reacted with each indicated protein (20 pmol). And the unwinding activity was assessed via gel electrophoresis and scanning on a Typhoon 9500 imager. Non-boiled reaction mixture (lane 1) and reaction mixture with MBP alone (lane 3) were used as negative controls, and boiled reaction mixture (lane 2) and reaction mixture with MBP-fusion EV71 2C (lane 5) were used as positive controls. (C) The unwinding activities at different MBP–VP35 or MBP-2C concentrations were plotted as the percentage of the released RNA from the total RNA helix substrate (Y-axis) at each protein concentration (X-axis). (D) MBP–VP35 (20 pmol) was reacted with the standard RNA helix substrate (0.1 pmol) in reaction mixture in the absence (lane 3) or presence (lane 4) of 5 mM ATP, or in the presence of 5 mM AMP–PNP (lane 5), followed by unwinding assay. (E) The RNA helix unwinding assay as described in (A) was performed in the presence of increasing concentrations of ATP. (F) The RNA helix unwinding assay was performed in the presence of each indicated NTP (5mM). For (C), error bars represent SD values from three separate experiments.