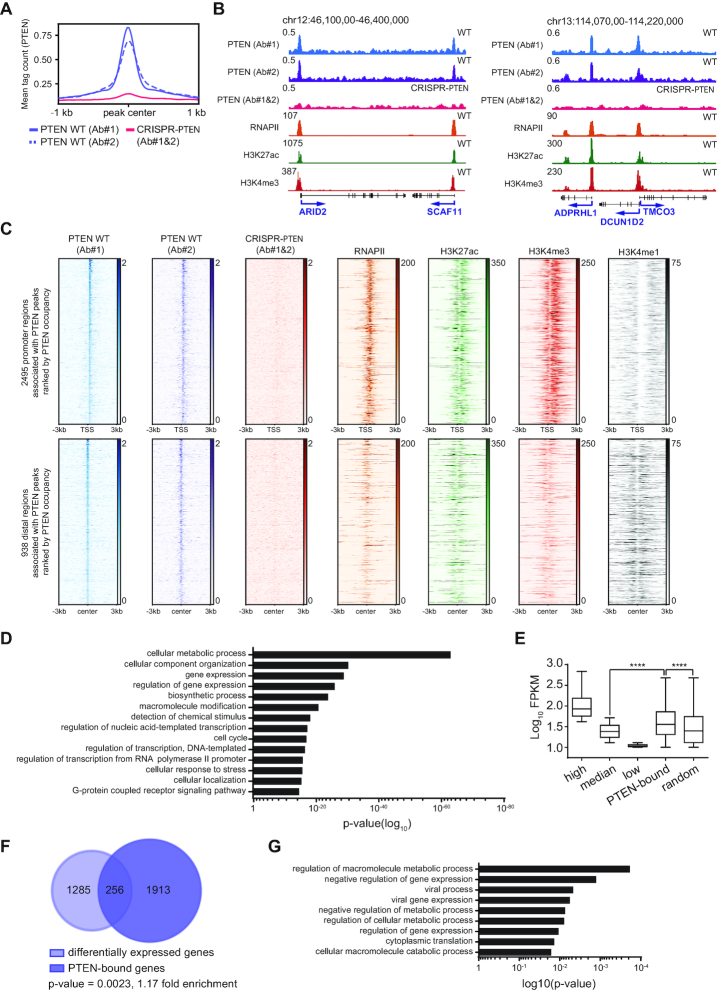
Figure 3.
PTEN binds to chromatin in promoter and putative enhancer regions. (A) PTEN ChIP-seq meta-profiles using two PTEN antibodies in PTEN WT (blue) and CRISPR-PTEN (red) HeLa cells. Data are centered and ±1 kb are shown. Plots represent average read counts per 50 bp bins. (B) Representative captures of the UCSC genome browser (GRCh37/hg19) showing PTEN, and publicly available RNAPII (44,45), H3K27ac, H3K4me1 and H3K4me3 (46,47) tracks in HeLa cells. The y-axis represents normalized read counts in reads per million. (C) Heatmaps of ChIP-seq reads for PTEN, and publicly available RNAPII (44,45), H3K27ac, H3K4me3 and H3K4me1 (46,47). Data are centered and ±3 kb are shown for (top) 2495 promoter regions and (bottom) 938 distal regions rank-ordered by PTEN occupancy. (D) GO enrichment analysis of PTEN-bound genes. p-values are given. (E) Log10FPKM of 3830 high, 7660 median, and 3830 low expressed genes, 2672 PTEN-bound and 2672 random genes in PTEN WT HeLa cells. Tukey boxplots with outliers omitted. Student's t-test, ****P ≤ 0.0001. (F) Hypergeometric gene set enrichment analysis to determine the overlap between differentially expressed and PTEN-bound genes. P-value and fold-enrichment are given. Calculation was based on 15 330 genes. (G) GO enrichment analysis of PTEN-bound genes that were significantly downregulated after PTEN loss. p-values are given.