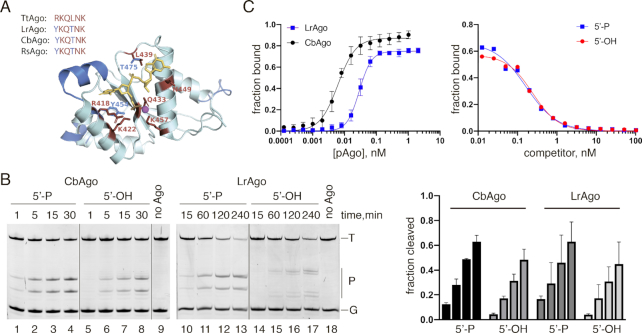
Figure 4.
CbAgo and LrAgo can utilize both 5′-phosphorylated and 5′-hydroxyl DNA guides at 37°C. (A) A 3D model of the LrAgo MID domain aligned to the structure of TtAgo in complex with gDNA (with bound Mg2+ ion; PDB: 3HO1). The model was built using the SWISS-MODEL portal. Amino acid residues of the conserved MID-domain motif (red; shown for various pAgos above the structure (4)) and Mg2+ ion (magenta) involved in interactions with the first two guide nucleotides (yellow) are highlighted. Elements of the secondary structure and amino residues specific to LrAgo are shown in blue. (B) Programmable ssDNA cleavage by CbAgo and LrAgo in the presence of either 5′-P or 5′-OH guides. The reactions were performed at the 5:2:1 pAgo:guide:target molar ratio for indicated time intervals. (C) (Left) Binding of 18 nt phosphorylated DNA guide by CbAgo and LrAgo. The fraction of bound DNA was plotted against protein concentration and fitted using the model of specific binding with the Hill slope. (Right) Analysis of gDNA binding by a competition assay. Radiolabeled gDNA (100 pM) was combined with increasing amounts of unlabeled 5′-P (blue) or 5′-OH (red) competitor, and incubated with LrAgo (100 pM). The fraction of bound DNA was plotted against competitor concentration and fitted using the one-site competitive binding model. Means and standard deviations from 3 independent experiments are shown.