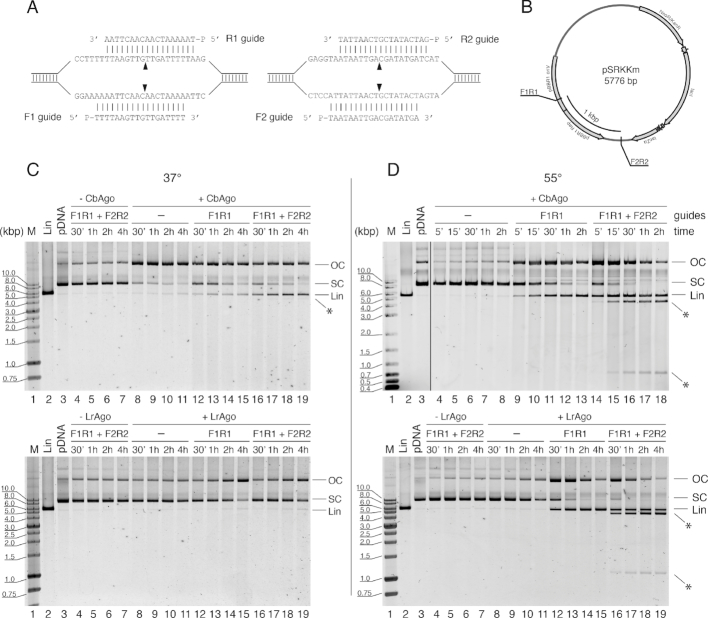
Figure 6.
CbAgo and LrAgo act as programmable DNA-nucleases for double-stranded DNA cleavage in vitro. (A) Target regions of the pSRKKm plasmid with two pairs of DNA guides (F1R1 and F2R2). Black triangles indicate the predicted cleavage sites. (B) Scheme of the pSRKKm plasmid with the positions of the two target regions. (C) Plasmid cleavage by CbAgo (top) and LrAgo (bottom) at 37°C. (D) Plasmid cleavage at 55°C. The reactions were performed with no guides, one pair of guides (F1R1) or two pairs of guides for indicated time intervals. Control reactions contained no pAgo proteins. Note that under the conditions of our assay, the supercoiled form of this plasmid migrated slower on the agarose gel than the linear plasmid, which identity was confirmed by comparison with the plasmid treated with a restriction endonuclease (KpnI, lane 2 in all panels). FR, forward and reverse guide DNAs; M, molecular weight marker; Lin, linearized plasmid; OC, open circular plasmid; SC, supercoiled plasmid; positions of specific cleavage products are indicated with asterisks.