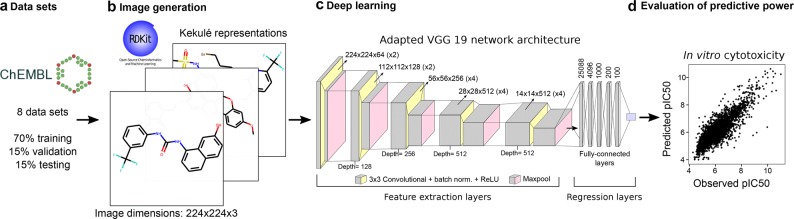
Fig. 1.
KekuleScope framework. a We collected and curated a total of 8 cytotoxicity data sets from ChEMBL version 23. b Compound Kekulé representations were generated for all compounds and used as input to the ConvNets. c We implemented extended versions of 4 commonly used architectures (e.g., VGG-19-bn shown in the figure) by including five additional fully-connected layers to predict pIC50 values on a continuous scale. d The generalization power of the ConvNets was assessed on the test set, and compared to RF models trained using Morgan fingerprints as covariates