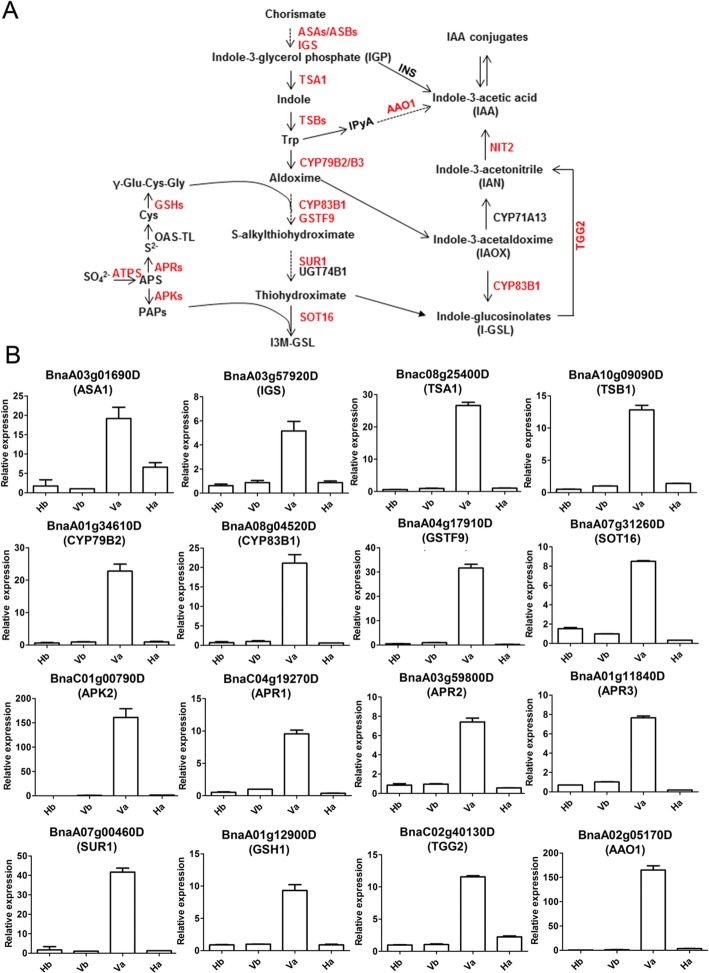
Fig. 3.
Validation of the DEGs involved in indole GLS-linked auxin biosynthesis via qRT-PCR. a The enriched metabolic pathway for the significant DEGs involved in secondary dormancy. The DEGs identified in the current study are marked in red. The genes in black were not detected in this study. The solid arrow indicates the direct promotion of the conversion, while the dotted arrow denotes the indirect promotion. b Verification of the enriched DEGs in (A) via qRT-PCR. Three independent biological replicates were tested and analysed. The relative expression levels of the DEGs were calculated via the 2-ΔΔCT method. The corresponding gene name is shown in parentheses, as annotated by BLASTN in Arabidopsis. BnaCAT1 was used as an internal control. Data are mean ± SD of three independent replicates