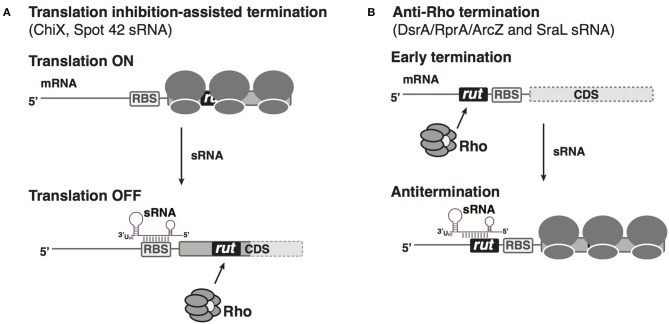
Figure 2.
sRNA-based negative or positive regulation of Rho-dependent transcription attenuation in bacteria. (A) Translation inhibition by sRNA assists Rho-dependent termination. When the accumulation of Salmonella ChiX or E. coli Spot 42 sRNA is low, the mRNAs chiPQ and galETKM are translated well. When ChiX or Spot42 sRNA is produced at high levels, under specific environmental conditions, sRNA annealing blocks the ribosome binding site of the corresponding mRNA target (chiP or galK, respectively); reduced translation allows Rho loading onto a cryptic rut site to terminate transcription inside the genes (dotted pale portion of box represents untranscribed portion of the mRNA). (B) Antagonization of Rho-dependent attenuation by sRNAs. In genes with long 5′ UTRs, rut sites can allow Rho termination, blocking expression of the downstream gene (dotted box represents untranscribed coding region). sRNAs binding within the 5′ UTR can block Rho access to the rut sites, thus acting as anti-terminators for the downstream CDS.