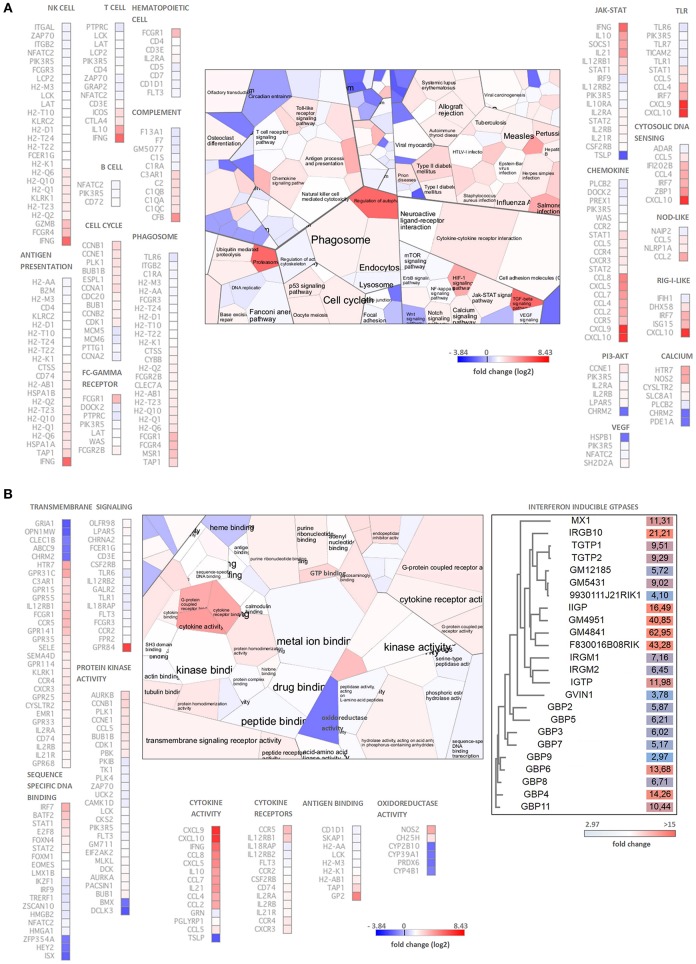
Figure 2.
Functional analysis of the C. muridarum infection altered genes by the Voronto method. (A) Analysis of significantly enriched KEGG pathways containing differentially expressed genes. (B) Gene Ontology analysis of significantly enriched molecular function terms containing differentially expressed genes. The differentially expressed IFN-inducible GTPases are grouped based on their amino acid sequences shown separately with their fold of up-regulation. Chlamydia induced inflammation, immunity and antichlamydial defense related Gene Ontology terms and KEGG pathways and the corresponding genes are shown separately. Cell colors of the Voronto diagrams show the mean fold change (log2) of the differentially expressed genes related to the particular Voronto cells. Both for the Voronto cells and the genes, the log2 fold changes are color coded according to the scales. Note that the colors of the up-regulated genes ranging from light blue to red, while the down-regulated genes are dark blue.