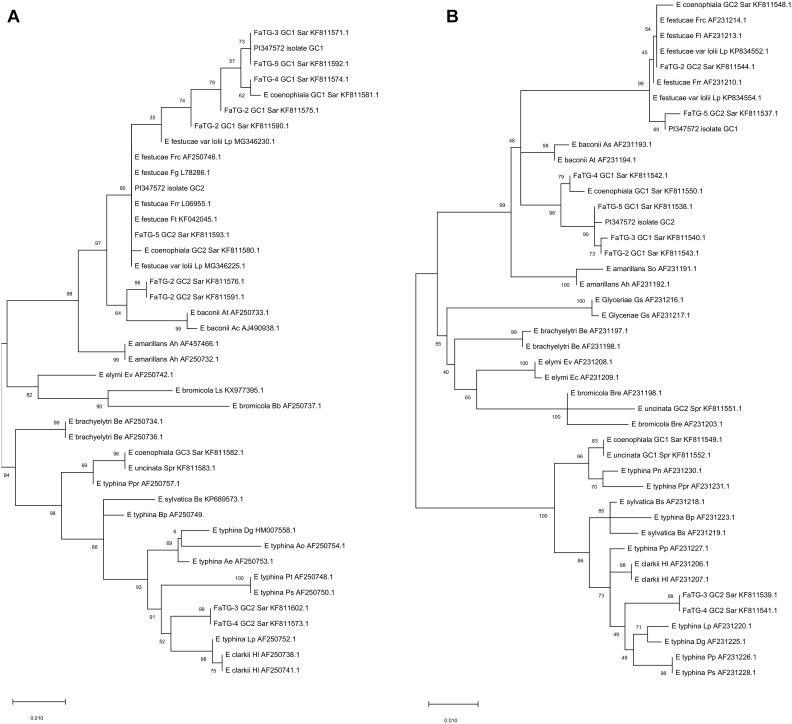
FIGURE 2.
Phylograms derived from maximum likelihood (ML) analysis and Tamura-Nei model of the partial sequence of tubB (A) and tefA (B) genes from representative haploid Epichloë species and hybrid species included in this study. The two copies obtained from S. arundinaceus var. glaucescens are labeled as “PI347572 isolate.” The tree is midpoint rooted and drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap values (1000 replicates) are shown next to the branches. GC stands for gene copy. GenBank accession numbers are provided for each sequence. Letters after each endophyte refer to host designations as follows: Lp, Lolium perenne; Dg, Dactylis glomerata; Ps, Poa sylvicola; Pp, Phleum pratense; Sar, Schedonorus arundinaceus; HI, Holcus lanatus; Bp, Brachypodium pinnatum; Bs, Brachypodium sylvaticum; Spr, Schedonorus pratensis; Pn, Poa nemoralis; Ppr, Poa pratensis; Gs, Glyceria striata; Be, Brachyelytrum erectum; Ev, Elymus virginicus; Ec, Elymus canadensis; Bre, Bromus erectus; So, Sphenopholis obtusata; Ah, Agrostis hiemalis; As, Agrostis stolonifera; At, Agrostis tennuis; Frr, Festuca rubra subsp. rubra; Fl, Festuca longifolia; Frc, Festuca rubra subsp. commutata; Fg, Schedonorus giganteus; Bb, Bromus benekenii; Ls, Leymus secalinus; Pt, Poa trivialis; Ao, Anthoxanthum odoratum.