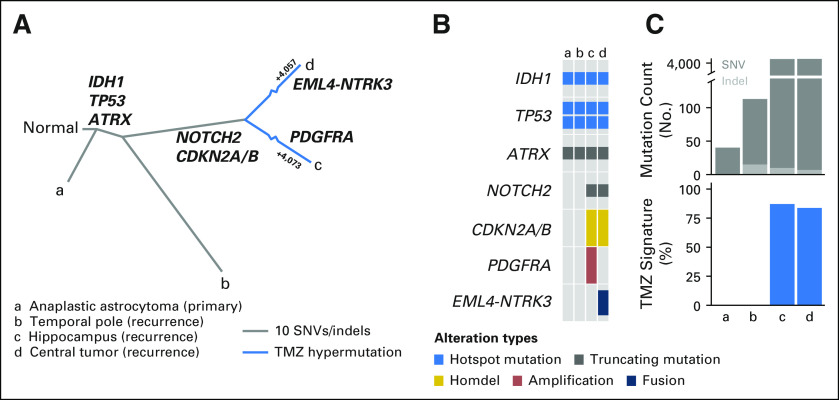
Fig 3.
Molecular characterization of initial and recurrent glioma. (A) Evolutionary relationships among sequenced tumor specimens inferred from somatic mutational data. Key functional alterations are shown for the branches in which they arose (bold). Among hypermutated specimens (c and d), branches are abbreviated and the number of temozolomide (TMX) -induced mutations in each lesion is specified. (B) Pattern of hallmark truncal mutations in IDH1, TP53, and ATRX across the initial tumor (a) and recurrent specimens (b-d) as well as private driver events in postprogression samples (c and d, as indicated). (C) Overall somatic mutational burden (top) of all tumor samples is shown, which indicates hypermutation in lesions c and d, but not in the initial pretreatment tumor or post-TMZ temporal pole tumor (b). At bottom is the fraction of mutations per sample attributable to the mutagenic effect of TMZ. SNV, single nucleotide variant.