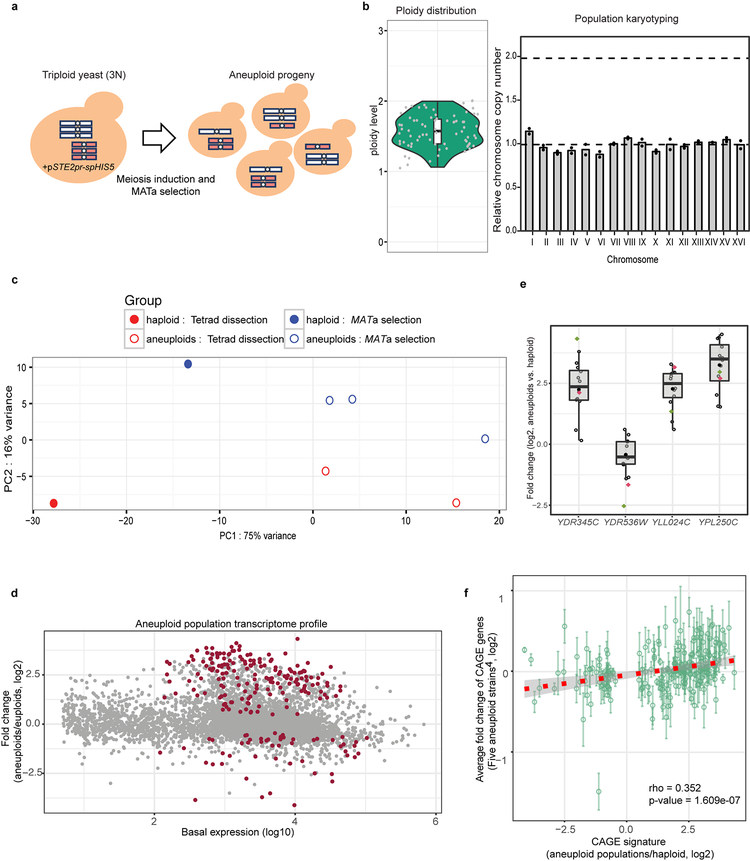
Extended Data Figure 1 |. Generation of heterogeneous aneuploid populations without chromosome copy number bias for transcriptomic analyses.
a. Generation of aneuploid cells through random meiotic segregation from a homozygous triploid (3N) yeast strain bearing a centromeric plasmid carrying the selection marker spHIS5 under the MATa-specific promoter STE2pr. Colored rods represent chromosomes. b. Left panel: DNA content of a cohort of random aneuploid spore colonies (each represented by a grey dot, n=75; Source data) produced through triploid meiosis analyzed by FACS; box-plot and violin-plot (defined as in Fig. 2 and Methods) show the distribution of ploidy levels. Right panel: 192 random aneuploid colonies produced by triploid meiosis were pooled and the resulting population subjected to qPCR-based karyotyping (representative results from two biological repeats). Y-axis represents the relative copy number (mean of the two arms) of each chromosome (dots) to the reference haploid yeast. There was no significant difference in copy number between each chromosome (One-way ANOVA, p-value = 0.998). c. Principal component analysis of RNA-seq results in different populations. PC1 (x-axis) shows apparent difference between haploid (n=2) and aneuploid (n=5) populations. PC2 (y-axis) reflects a small difference in the two methods of generation of aneuploid populations. Each dot represents one population. d. Gene expression in heterogeneous aneuploid populations (n=5) relative to haploid populations (n=2) in MA plot. X-axis represents basal mean expression in log10 scale; Y-axis represents differential expression changes between aneuploid and euploid populations in log2 scale. P-values calculated on the Wald statistic were corrected for multiple comparisons (Benjamini-Hochberg) and the resulting FDR was further corrected for variance underestimation using an empirical null model. CAGE genes were identified as genes with final FDR < 0.05 (see details in Supplementary Methods; exact p-values in Supplementary Table 1). Each dot represents one gene, and 222 significantly differentially expressed (CAGE) genes common to all five aneuploid cell populations are labeled in dark red. e. RT-qPCR validations of four significantly differentially expressed genes from the RNA-seq analysis in a heterogeneous aneuploid population (n=1) and randomly picked individual aneuploid clones (n=12). Black circles: individual aneuploids, red dot: a heterogeneous aneuploid population, and green dot: aneuploid populations in RNA-seq (d). For individual aneuploid colonies, expression change of each gene was normalized by the gene copy number (determined by qPCR), and then normalized to that of a non-CAGE gene (ACT1). Average gene expression changes in the population and in the individual aneuploid clones followed similar trends (up or down regulated) as heterogeneous populations in RNA-seq. Boxplot representation as in Fig. 2 for measurements from the 12 aneuploid clones. f. Correlation analysis of expression changes of the genes implicated in the CAGE signature showing positive correlation between aneuploid populations and stable aneuploid strains (n=5)5 (Spearman’s rank correlation). Green circles represent mean expression changes of genes within the CAGE signature (213 in common in the transcriptomic data between aneuploid populations and the individual strains); error bars: SEM. Only eight genes across all five strains appear to show opposite expression trend to CAGE. The trendline was fitted by linear regression (red dashed line) with grey ribbon (95% CI). Significant positive correlation was observed between the average gene expression changes in the five aneuploid strains and CAGE observed with the heterogeneous aneuploid population subjected to RNAseq (p-value = 1.609e-07).