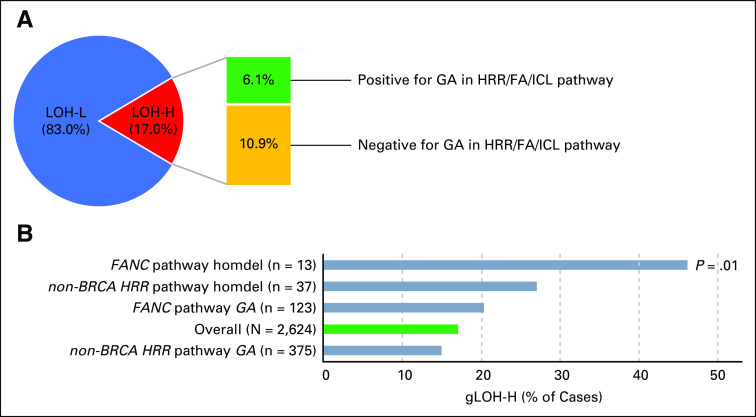
FIG A4.
(A) Genome-wide loss of heterozygosity (gLOH) score was assessable for 2,624 cases, and cases with 14% or more gLOH were designated LOH-high (LOH-H). All other cases were LOH-low (LOH-L). LOH-H cases were assessed for the presence (green) or absence (yellow) of a genomic alteration (GA) in the homologous recombination repair (HRR) or Fanconi anemia/interstrand crosslink repair (FA/ICL) pathway. (B) Genes were grouped together into HRR pathway (excluding non-BRCA1/2) or FA/ICL pathway (FANC); genes were categorized as in Figure 2A. All GAs or only homozygous deletions (homdel) were assessed for LOH-H and compared with the overall data set (green). Each subset was individually compared with the overall data set, and unadjusted P values are shown (Fisher’s exact test [two-tailed]).