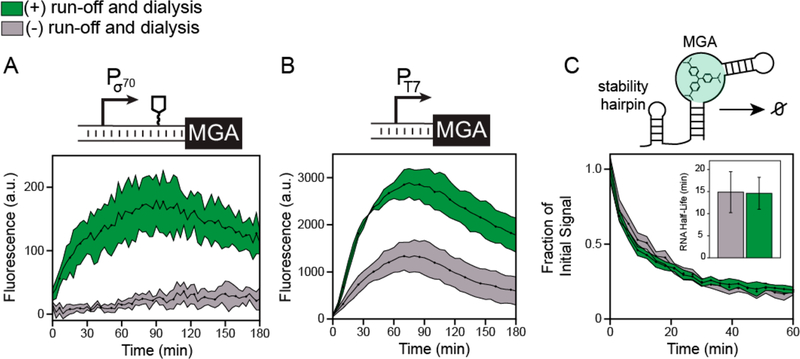
Figure 3.
Postlysis processing enhances the transcription rate but does not affect RNA degradation rates in CFE systems. (A) In vitro transcription of the malachite green RNA aptamer (MGA) from a strong E. coli σ70 promoter in a cell-free gene expression reaction containing the malachite green dye using either processed (green) or nonprocessed (gray) extracts. The construct encoded a 5’ stability hairpin before the MGA DNA sequence to enhance fluorescence observation. (B) In vitro transcription of the MGA construct from a T7 promoter and no 5’ stability hairpin. Kinetic data in (A) and (B) represent the average of three technical replicates, with shaded region representing plus-minus one standard deviation of the mean. (C) Characterization of RNA degradation rates. The malachite green RNA aptamer was purified with the 5’ stability hairpin, mixed with the malachite green dye and 10% extract by volume, and the decay in fluorescence over time was measured to quantify RNA degradation rates (kdegradation)· Inset: the RNA half-life (ln(2)/kdegradation) was estimated by fitting an exponential decay function to the fluorescence kinetics over the first 30 min to fit kdegradation. Errors in half-lives are propagated from three independent measurements and reported as standard deviation.