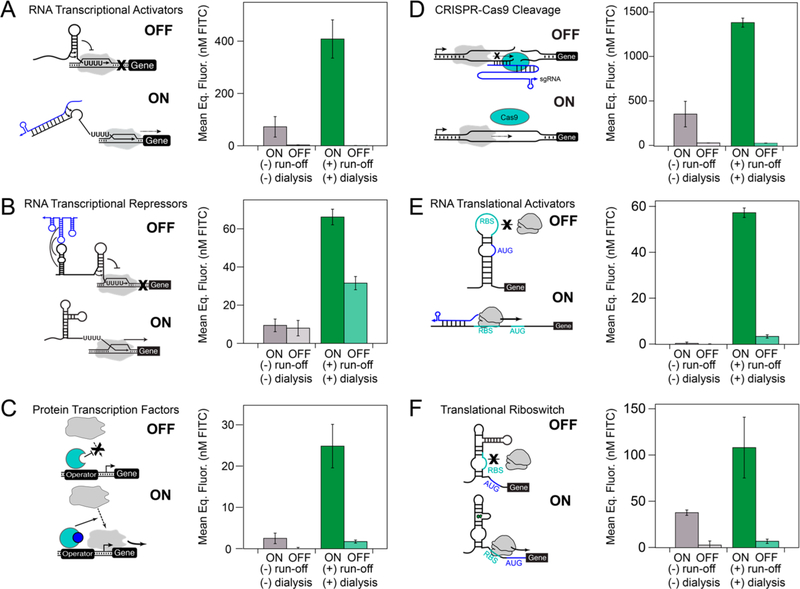
Figure 5.
Processed extracts enable cell-free expression of a wide array of genetic circuitry. Postlysis processing enables significantly higher ON states, including activation in some cases, for (A) small RNA transcriptional activators (STARs), (B) RNA transcriptional repressors, (C) LuxR, the ;Y-acyl homoserine lactone (AHL)-inducible σ70 transcription factor, (D) CRISPR-Cas9 cleavage, (E) toehold switch RNA translation activators, and (F) translational riboswitches. Each experiment was done in technical triplicate in either a postlysis processed (green) or nonprocessed (gray) extract for 8 h at 30 °C in a plate reader monitoring sfGFP production. The total plasmid DNA concentration was held constant in each reaction between the ON and OFF states of each circuit to remove variation caused by unwanted cell-free transcription and translation of the antibiotic resistance genes encoded on each plasmid. Complete reaction conditions including plasmid and inducer concentrations and optimal magnesium levels are included in Table S1. Kinetics of gene expression trajectories for each circuit are presented in Figure S7. Error bars represent the standard deviation of the mean for three technical replicates drawn from a single batch of each extract.