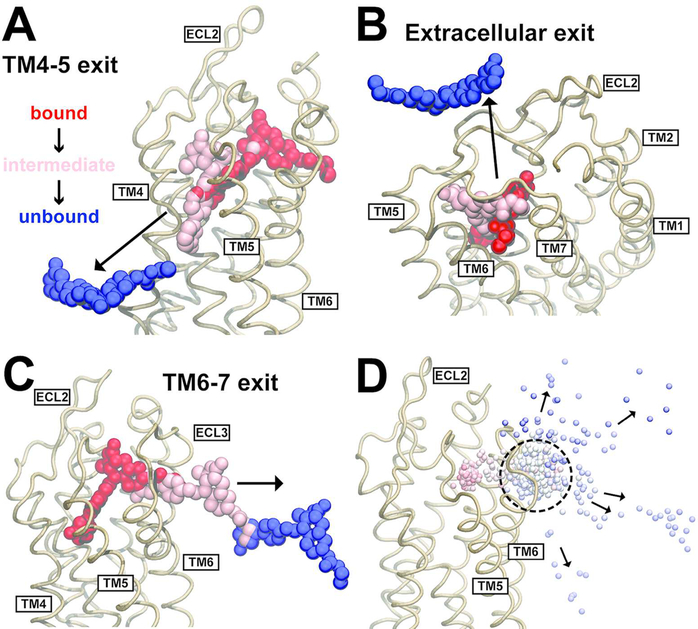
Figure 2.
Ligand dissociation trajectories observed in TAMD simulations of vorapaxar dissociating from PAR1. (A) Ligand dissociation pathway between TM4 and TM5, observed in three simulations. (B) Ligand dissociation pathway into the extracellular solvent, observed in five simulations. (C) Ligand dissociation pathway between TM6 and TM7, observed in five simulations. In panels A–C, the PAR1 backbone is shown as light brown ribbons and vorapaxar atoms are shown as spheres. The crystallographic pose of vorapaxar is colored red, as well as representative poses from intermediate (pink) and unbound states (blue) observed in the simulations. (D) Overlay of the COM of vorapaxar (small spheres) during dissociation from PAR1 (light brown ribbons) between TM6 and TM7 in five different simulations. Simulation time progresses from red to white to blue in each simulation, and the time step is 180 ps. The metastable state is indicated by a dashed circle. The direction of vorapaxar motion is indicated by arrows.