Abstract
Cytoplasmic microinjection (CI) of the CRISPR/Cas9 system enabled the induction of site-specific mutations in porcine zygotes and resulting pigs. However, mosaicism is a serious problem for genetically modified pigs. In the present study, we investigated suitable timing and concentration of CRISPR/Cas9 components for introduction into oocytes/zygotes by CI, to reduce mosaicism in the resulting blastocysts. First, we introduced 20 ng/μl of Cas9 protein and guide RNA (gRNA), targeting the α-1,3-galactosyltransferase (GalT) gene in oocytes before in vitro fertilization (IVF), in zygotes after IVF, or in oocytes/zygotes before and after IVF, twice. CI treatment had no detrimental effects on blastocyst formation rates. The highest value of the rate of mutant blastocysts was observed in zygotes injected after IVF. Next, we injected Cas9 protein and gRNA into zygotes after IVF at a concentration of 20 ng/μl each (20 ng/μl group) or 100 ng/μl each (100 ng/μl group). The ratio of the number of blastocysts that carried mutations to the total number of blastocysts examined in the 100 ng/μl group was significantly higher (P < 0.05) than that in the 20 ng/μl group. Although no blastocysts from the 20 ng/μl group carried a biallelic mutation, 16.7% of blastocysts from the 100 ng/μl group carried a biallelic mutation. In conclusion, increasing the concentration of Cas9 protein and gRNA is effective in generating biallelic mutant blastocysts. To reduce mosaicism, however, further optimization of the timing of CI, and the concentration of CRISPR/Cas9 components, is needed.
Keywords: CRISPR/Cas9, Cytoplasmic microinjection, Gene editing, Mosaicism, Porcine zygote
Pigs are considered excellent laboratory animals because of their close similarity to humans, both anatomically and physiologically. Moreover, pigs are expected to contribute to research in human medicine, including regenerative medicine, as human disease models and as suitable organ donors for xenotransplantation achieved by the genetic modification of pigs. The first genetically modified pigs were generated by embryo transfer of mutant embryos modified by microinjection of exogenous DNA into the pronucleus of zygotes [1]. However, the efficiency of introducing mutations was quite low because genetic modification by pronuclear microinjection of exogenous DNA relies on random insertion of genes [2]. After the establishment of somatic cell nuclear transfer (SCNT) in pigs, SCNT is now a main technique used to generate genetically modified pigs from genetically modified donor somatic cells due to the greater efficiencies of preparing donor cells carrying site-specific changes and a wide range of modifications for knockout and knockin [3, 4]. However, SCNT requires sophisticated techniques, and successful SCNT depends on the operator’s skills. Birth defects, abortion, and early postnatal death are also major problems associated with SCNT, followed by embryo transfer of reconstructed embryos [5]. Thus, the efficiency of generating live genetically modified pigs by SCNT has been low [3].
Gene editing with site-specific nucleases, such as clustered regularly interspaced short palindromic repeat (CRISPR) / CRISPR-associated nuclease 9 (Cas9) [6], has enabled the introduction of site-specific mutations in animal cells. Cytoplasmic microinjection (CI) of CRISPR/Cas9 components, Cas9 mRNA/protein and guide RNA (gRNA), also enabled the induction of site-specific mutations in zygotes and embryos [3]. However, the genotypes of mutant embryos from zygotes injected with CRISPR/Cas9 components often exhibit mosaic patterns. These mutant embryos are composed of several types of cells with different mutations [7,8,9,10], which have often been observed after pronuclear microinjection of foreign DNA [11]. The occurrence of genetic mosaicism in resultant pigs is a serious problem because mosaicism, including wild-type (WT) cells, complicates phenotypic analysis. Although it is necessary to produce a non-mosaic F1 generation from the mosaic mutants to analyze phenotypes clearly, the production of the next generation is time-consuming and labor intensive, particularly in large animals because of the much longer pregnancy period than that in mice.
Mosaicism is predicted to appear due to the occurrence of gene editing after the first replication of the zygotic genome. To generate non-mosaic mutants, including biallelic mutants, the CRISPR/Cas9 system needs to be functional prior to the first genome replication. Therefore, one of the important factors to control mosaicism seems to be the timing of introducing CRISPR/Cas9 components into 1-cell zygotes. However, there is limited information regarding the timing of injection of the CRISPR/Cas9 components into porcine zygotes. On the other hand, it has been reported that the concentration of Cas9 mRNA and gRNA introduced into mouse zygotes by electroporation influences the efficiency of gene editing [12]. In the case of gene editing by CI using the Cas9 protein, the optimization of functional concentrations of Cas9 protein and gRNA is also required to reduce mosaicism. However, few researchers have evaluated the effects of these factors on gene editing of porcine zygotes using the CRISPR/Cas9 system in order to reduce mosaicism in the resulting blastocysts.
In this study, we examined the suitable timing of introducing Cas9 protein with gRNA into in vitro-fertilized zygotes by CI. We also evaluated the concentration of Cas9 protein and gRNA for efficient gene editing by CI.
Materials and Methods
Oocyte collection and in vitro maturation
Porcine ovaries were obtained from approximately 6-month-old gilts from a local slaughterhouse and were transported in physiological saline within 1 h to the laboratory at 30°C. Ovaries were washed three times with modified phosphate-buffered saline (m-PBS; Nihonzenyaku, Fukushima, Japan) supplemented with 100 IU/ml penicillin G potassium (Meiji, Tokyo, Japan) and 0.1 mg/ml streptomycin sulfate (Meiji). The follicles of the ovarian surface were sliced on a sterilized dish using a surgical blade, and cumulus-oocyte complexes (COCs) were visualized and collected under a stereomicroscope. Only COCs with a uniformly dark-pigmented ooplasm and intact cumulus cell masses were collected. Approximately 50 COCs were then cultured in 500 µl of maturation medium consisting of tissue culture medium 199 with Earle’s salts (TCM 199; Invitrogen, Carlsbad, CA, USA) supplemented with 10% (v/v) porcine follicular fluid, 0.6 mM cysteine (Sigma-Aldrich, St. Louis, MO, USA), 50 µM β-mercaptoethanol (Wako Pure Chemical Industries, Osaka, Japan), 50 µM sodium pyruvate (Sigma-Aldrich), 2 mg/ml D-sorbitol (Wako Pure Chemical Industries), 10 IU/ml equine chorionic gonadotropin (Kyoritu Seiyaku, Tokyo, Japan), 10 IU/ml human chorionic gonadotropin (Kyoritu Seiyaku), and 50 µg/ml gentamicin (Sigma-Aldrich), then covered with mineral oil (Sigma-Aldrich) for 22 h in 4-well dishes (Nunc A/S, Roskilde, Denmark). Subsequently, the COCs were transferred into maturation medium without hormone supplementation and cultured for an additional 22 h. The incubation of COCs was conducted at 39°C in a humidified incubator containing 5% CO2.
In vitro fertilization
In vitro fertilization (IVF) was performed according to methods described previously [13]. Frozen-thawed ejaculated spermatozoa were transferred into 5 ml of fertilization medium (PFM; Research Institute for the Functional Peptides, Yamagata, Japan) and washed by centrifugation at 500 × g for 5 min. The pelleted spermatozoa were resuspended in fertilization medium and adjusted to 1 × 106 cells/ml. Next, approximately 50 oocytes were transferred to 500 µl of sperm-containing fertilization medium, covered with mineral oil in 4-well dishes, and co-incubated for 5 h at 39°C in a humidified incubator containing 5% CO2, 5% O2, and 90% N2.
Cytoplasmic microinjection
The matured oocytes and presumptive zygotes were subjected to CI of CRISPR/Cas9 components. Microinjection was performed in a 20 μl drop of PZM-5 (Research Institute for the Functional Peptides) covered by mineral oil. Opti-MEM I solution containing Cas9 protein (Guide-it™ Recombinant Cas9; Takara Bio, Shiga, Japan) and gRNA (Alt-R™ CRISPR crRNAs and tracrRNA, chemically modified and length-optimized variants of native guide RNAs purchased from IDT (Integrated DNA Technologies, Coralville, IA, USA)) targeted exon 1 of the α-1,3-galactosyltransferase (GalT) gene, which encodes a specific antigen on the surface of pig cells, which is a major obstacle in pig-to-human xenotransplantation [14]. The target sequence: (5′-TGTTTTGGGAATACATCAAC-3′) was loaded into an injection pipette (Femtotips II, Eppendorf, Hamburg, Germany). Oocytes/zygotes were immobilized with a holding pipette, and the tip of the injection pipette was inserted into the cytoplasm through the zona pellucida and the cell membrane at the 3 o’clock position. Subsequently, opti-MEM I solution containing Cas9 protein and gRNA was injected into the cytoplasm by air pressure (injection pressure, 100–200 hectopascals (hPa); compensation pressure, 10–15 hPa; and injection time, 0.15 sec) using a microinjector (FemtoJet 4i; Eppendorf). We confirmed the success of CI by observing slight swelling of the oocyte/zygote cytoplasm.
In vitro culture
The zygotes were subsequently transferred to PZM-5. Approximately 50 zygotes were cultured continuously in 500 µl of PZM-5 covered with mineral oil for three days in 4-well dishes. Then, the zygotes were cultured in 500 µl of porcine blastocyst medium (PBM; Research Institute for the Functional Peptides) covered with mineral oil for four days in 4-well dishes. The incubation of zygotes and embryos was conducted at 39°C in a humidified incubator containing 5% CO2, 5% O2, and 90% N2. To evaluate the gene editing efficiency, blastocysts from microinjected zygotes were collected on day 7 (day 0 = insemination) and subjected to gene analysis, as described below.
Analysis of targeted genes after CI
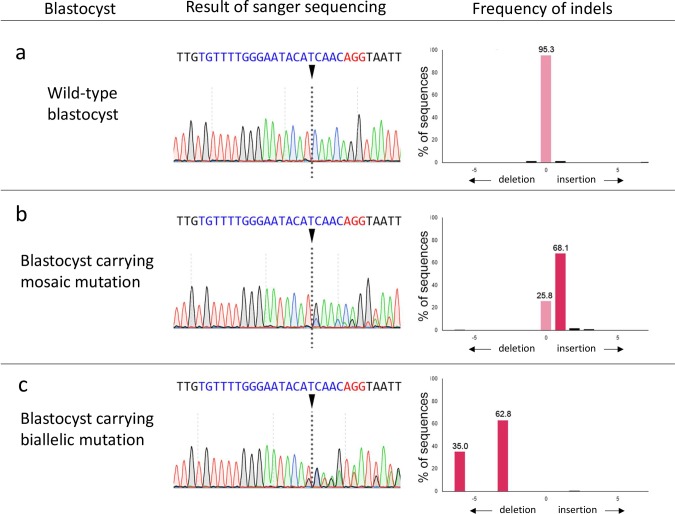
Genomic DNA was isolated from blastocysts by boiling them in a 50 mM NaOH solution. After neutralization, the genomic regions flanking the gRNA target sequences were PCR-amplified using specific primers targeting exon 1 of the GalT gene, including the target site of the gRNA: 5ʹ-AGTCAGGATGCTTCCCCTTT-3ʹ (forward) and 5ʹ-AAGCTGGTGACTTGGCTGAT-3ʹ (reverse). Forward and reverse PCR primers were designed 341 bp upstream and 83 bp downstream of the anticipated gRNA cut site, respectively. PCR products from blastocysts were extracted by agarose gel electrophoresis. The targeted genomic regions of the PCR products were directly sequenced. Sanger sequencing was performed using BigDye Terminator Cycle Sequencing Kit ver. 3.1 (Thermo Fisher Scientific, Waltham, MA, USA) and an ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) platform. The TIDE bioinformatics package [15] was used to determine the gene editing efficiency in blastocysts. Blastocysts that carried no WT sequences were classified as having biallelic mutations. Blastocysts that carried one or more types of mutation with the WT sequence were classified as mosaics. Blastocysts that only carried the WT sequence were classified as WT (Fig. 1). The mutation efficiency was defined as the ratio of the number of blastocysts that carried mutations to the total number of blastocysts examined.
Fig. 1.
Representative results of Sanger sequencing of blastocysts formed after cytoplasmic microinjection with Cas9 protein and GalT gRNA, and the frequencies of indels decomposed from Sanger sequence data by TIDE analysis. (a) Analysis of wild-type blastocyst. (b) Analysis of a blastocyst carrying mosaic mutation. The Sanger sequence traces consist of a mixture of wild-type and 1 bp insertion sequence, which yields a composite sequence trace after the break site. TIDE analysis decomposed the Sanger sequence data, and indels and their frequencies are indicated. (c) Analysis of blastocyst carrying biallelic mutation. The Sanger sequence traces of a blastocyst carrying biallelic mutation consist of a composite sequence trace without wild-type sequence. Arrowheads indicate the Cas9 cleavage sites. Nucleotides in blue color indicate target sequences, and nucleotides in red color indicate protospacer adjacent motif (PAM) sequences.
Experimental design
Experiment 1: To confirm the optimum timing of CI for efficient gene editing, we performed CI in oocytes before IVF, in zygotes after IVF, and in oocytes and zygotes before and after IVF, twice. Briefly, matured oocytes were denuded from cumulus cells by treatment with 150 IU/ml hyaluronidase (Sigma-Aldrich) in Medium 199, with gentle pipetting. The denuded oocytes were subjected to IVF and CI. The oocytes/zygotes were injected with 20 ng/μl each of Cas9 protein and gRNA, targeting the GalT gene, 2 h before the start of IVF (CI-IVF group), 6 h after the start of IVF (IVF-CI group), or 2 h before the start of IVF and 6 h after the start of IVF (CI-IVF-CI group), twice. After IVF, the putative zygotes were denuded from the attached spermatozoa by gentle pipetting. After IVF and CI, the zygotes were cultured for seven days, as described above. As a control, some zygotes without CI treatment before and after IVF were cultured for the same number of days. The resultant blastocysts after CI treatment were collected and subjected to analysis of gene editing efficiency, as described above.
Experiment 2: The effect of Cas9 and gRNA concentration on gene editing efficiency was examined. After IVF, putative zygotes were injected with 20 ng/μl each or 100 ng/μl each of Cas9 protein and gRNA 6 h after the start of IVF. After in vitro culture for seven days, the resulting blastocysts were collected and subjected to analysis of gene editing efficiency, as described above.
Statistical analysis
All percentage data were subjected to arcsine transformation before performing analysis of variance (ANOVA). The transformed data were tested by ANOVA, followed by Fisher’s protected least significant difference (LSD) test, using StatView software (Abacus Concepts, Berkeley, CA, USA). The percentages of mutations in the total number of blastocysts were analyzed by chi-squared analysis. Differences with a probability value (P) of 0.05 or less were considered statistically significant.
Results
Experiment 1
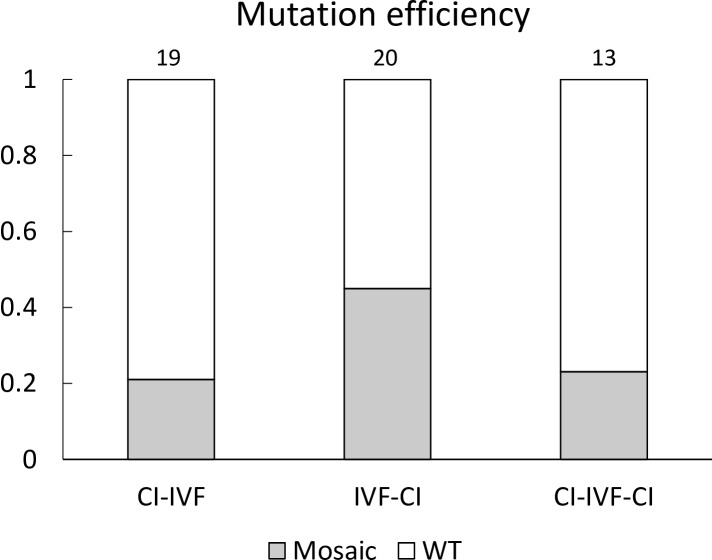
As shown in Table 1, the cleavage rates of embryos after CI-IVF-CI treatment were significantly lower (P < 0.05) than those of control embryos. However, CI treatment had no effect on blastocyst formation rates. When the rates of blastocysts carrying mutations were compared between the experimental groups with CI treatment, the highest value was observed in the IVF-CI group (Fig. 2). Various types of insertion/deletion (indel) mutations, ranging from 6-bp deletions to 3-bp insertions, were detected in mutant blastocysts. There were no biallelic mutant blastocysts in any of the treatment groups.
Table 1. Effects of microinjection treatment on the development of porcine zygotes *.
| Experimental group ** | Number of embryos examined | Number of embryos |
|
|---|---|---|---|
| cleaved (%) | developed to blastocysts (%) | ||
| Control | 225 | 203 (90.2 ± 1.0) a | 22 (9.5 ± 1.5) |
| CI-IVF | 238 | 187 (77.9 ± 3.3) ab | 21 (8.2 ± 2.1) |
| IVF-CI | 269 | 200 (74.5 ± 3.5) ab | 17 (6.4 ± 1.2) |
| CI-IVF-CI | 234 | 137 (57.3 ± 14.6) b | 14 (5.9 ± 1.6) |
* Five replicate trials were carried out. Percentages are expressed as the mean ± SEM. ** CI: cytoplasmic microinjection, IVF: in vitro fertilization, CI-IVF: CI at 2 h before the start of IVF, IVF-CI: CI at 6 h after the start of IVF, CI-IVF-CI: CI at 2 h before the start of IVF, and 6 h after the start of IVF, twice. a, b Values with different superscripts in the same column differ significantly (P < 0.05).
Fig. 2.
Mutation efficiency of blastocysts resulting from zygotes injected with Cas9 protein and GalT gRNA at various time points. The mutation efficiency indicates the ratio of the number of blastocysts that carried mutations to the total number of blastocysts examined. Numbers above the bars indicate total number of blastocysts examined. Mosaic: mosaic mutant, WT: wild-type, CI: cytoplasmic microinjection, IVF: in vitro fertilization, CI-IVF: CI at 2 h before the start of IVF, IVF-CI: CI at 6 h after the start of IVF, CI-IVF-CI: CI at 2 h before the start of IVF and at 6 h after the start of IVF, twice.
Experiment 2
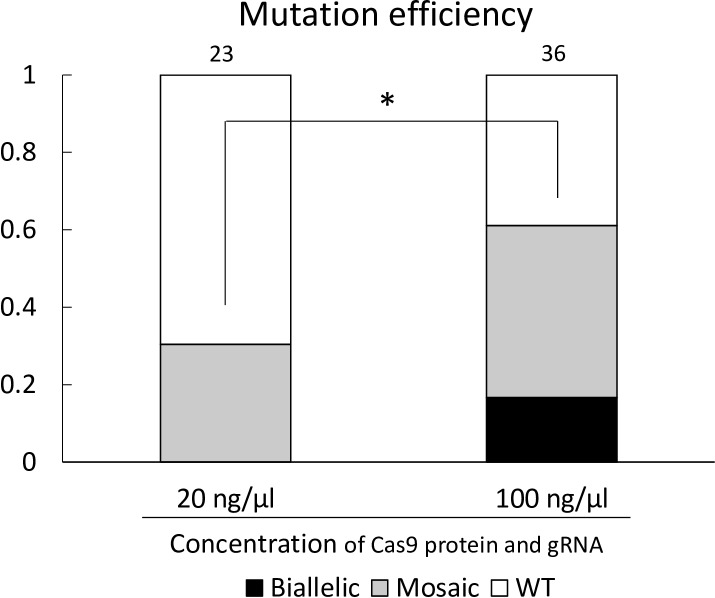
The cleavage rates of the Cas9 protein and gRNA injected group (74.0% and 70.9%, for 20 and 100 ng/μl each of Cas9 protein and gRNA injected group, respectively) were statistically similar to that of the control group (85.5%). Moreover, the blastocyst formation rates of the Cas9 protein and gRNA injected group (11.9% and 18.0%, for 20 and 100 ng/μl each of Cas9 protein and gRNA injected group, respectively) were also statistically similar to that of the control group (19.4%). The ratio of the number of blastocysts that carried mutations to the total number of examined blastocysts in the 100 ng/μl each of Cas9 protein and gRNA injected group (61.1%) was significantly higher (P < 0.05) than that in the 20 ng/μl each of Cas9 protein and gRNA injected group (30.4%) (Fig. 3). Although there were no biallelic mutant blastocysts in the 20 ng/μl each of Cas9 protein and gRNA injected group, 16.7% of blastocysts injected with 100 ng/μl each of Cas9 protein and gRNA carried biallelic mutations. Various types of indel mutations, including 8 bp deletions to 9 bp insertions, were detected in mutant blastocysts.
Fig. 3.
Mutation efficiency in blastocysts from zygotes injected with 20 ng/µl each or 100 ng/µl each of Cas9 protein and gRNA. The mutation efficiency indicates the ratio of the number of blastocysts that carried mutations to the total number of blastocysts examined. Numbers above the bars indicate total number of blastocysts examined. Biallelic; biallelic mutant, Mosaic: mosaic mutant, WT: wild-type. * Chi-squared analysis showed a significant difference between the mutation efficiency of blastocysts from zygotes injected with 20 ng/µl each of Cas9 protein and gRNA and those injected with 100 ng/µl each of Cas9 protein and gRNA (P < 0.05).
Discussion
In the present study, we found that performing CI in zygotes after IVF may be more beneficial than performing CI in oocytes before IVF as the former could lead to a higher number of blastocysts carrying mutations. Moreover, when the concentration of Cas9 protein and gRNA was increased to 100 ng/μl, the mutation efficiency was significantly increased, and some blastocysts successfully carried biallelic mutations.
In the first experiment, we found that when CI was performed at 6 h after the start of IVF, the highest value of the rate of blastocysts carrying mutations was observed. Therefore, we adopted performing CI at 6 h after the start of IVF as the suitable time point for introducing the CRISPR/Cas9 components in our system. A previous study using parthenogenetic embryos showed that microinjection of CRISPR/Cas9-related mRNA at 6 h after electrical activation to induce parthenogenesis increased gene editing efficiency, compared to that immediately after electrical activation [9]. The authors hypothesized that the formation of the pronucleus may correlate with increased mRNA translation, resulting in higher gene editing efficiency. On the other hand, the delayed introduction of the CRISPR/Cas9 system also causes a reduction of biallelic mutations [16]. CRISPR/Cas9-mediated mutation efficiency was evaluated using parthenogenetic embryos that were microinjected with Cas9 mRNA and gRNA at 3, 8, or 18 h after activation treatment, and the rates of biallelic mutant embryos microinjected at 3 and 8 h after activation were approximately four and five times higher than those at 18 h [16]. In pigs, it has been reported that male pronucleus formation and DNA replication begin at 8–9 h, and 9.7 h after intracytoplasmic sperm injection, respectively [17, 18]. Another study in in vitro fertilized porcine zygotes has indicated that male and female pronucleus formation, and fusion of two pronuclei, begin at 6–10 h, 8 h, and 18 h after fertilization, respectively [19]. At 18 h after activation, DNA replication may have commenced; thus, the frequency of mosaicism seems to increase. Although there are some differences between their studies and our study (e.g., we used in vitro fertilized zygotes and Cas9 protein), our present results support these previous studies, which indicate that the timing of introduction of the CRISPR/Cas9 system is an important factor modulating the frequency of mosaicism in embryos.
In the case of using mRNA for CRISPR/Cas9-mediated gene editing of zygotes, the delay in mRNA expression results in gene editing after the first replication of the zygotic genome, which induces mosaicism [7]. In the present study, we used Cas9 protein to examine the timing of CI because injection of Cas9 protein was considered to exhibit more rapid and efficient gene editing than injection of Cas9 mRNA with gRNA [20,21,22]. The mutation efficiency of the resulting blastocysts from oocytes injected before IVF was statistically similar to that from zygotes injected after IVF, but exhibited low values. In sheep, Cas9 mRNA and gRNA microinjection in MII oocytes resulted in improved mutation efficiency when compared to zygote microinjection [23]. The CRISPR/Cas9 activity introduced in oocytes before IVF may be different, depending on the species. Moreover, in human cells, the relative abundance of Cas9 protein delivered by transfection was greatly diminished by 24 h [20]. Therefore, our results also indicate that the stability of CRISPR/Cas9 components in cells is different from that in oocytes/embryos, and that CRISPR/Cas9 activity introduced into porcine oocytes before IVF might be decreased during the insemination process.
In the present study, some oocytes/zygotes underwent CI before and after IVF, twice (CI-IVF-CI group); however, the rate of gene editing in the resultant blastocysts was low. We observed that CI-IVF-CI treatment reduced the cleavage rate of oocytes/zygotes, likely indicating mechanical damage caused by the necessary micromanipulation procedures. Moreover, some studies observed cytotoxicity with the introduction of CRISPR/Cas9 components and the micromanipulation media [10, 24]. Our results indicate that the low efficiency of gene editing in the CI-IVF-CI group might result from mechanical damage and toxicity of the CRISPR/Cas9 system, after two rounds of CI with the micromanipulation media.
In the second experiment, we found that CI with a higher concentration of Cas9 protein and gRNA accelerated mutation efficiency and increased biallelic mutations in the resulting blastocysts. These results indicate that the gene editing efficiency of the CRISPR/Cas9 system appears to be proportional to the concentration of Cas9 protein and gRNA. To date, previous studies have used various concentrations of Cas9 mRNA, Cas9 protein, and gRNA at functional concentrations for microinjection in pigs, e.g., 20 ng/µl Cas9 mRNA and 10 ng/µl gRNA [25, 26], 125 ng/µl Cas9 mRNA and 12.5 ng/µl gRNA [16, 27], 25 ng/μl Cas9 protein and two kinds of 12.5 ng/μl gRNAs [24], 25 ng/μl each of Cas9 protein and gRNA [28]. However, comparative studies of the concentration of CRISPR/Cas9 components are few in number. Genetic mosaicism in resulting pigs is an obstacle for phenotypic analysis. On the other hand, to analyze the function of lethal genes, inducing intended mosaicism can be an effective strategy. In mice, chimeric founder generations with embryonic stem cells carrying biallelic mutations of a lethal gene suppressed the lethal phenotype and the animals survived to adulthood [29]. Generally, a number of genetically homogenous pups, including mono- and biallelic mutants, are needed to analyze gene function. Depending on the induced mosaic pattern and degree of mosaicism, the mosaic mutants are also expected to survive until sexual maturity, even if the targeted gene is lethal, which enables the generation of genetically homogenous F1 piglets. The mutation efficiency can be regulated by the control of the concentration of CRISPR/Cas9 components. Our study provides useful information for regulation of mosaicism in pigs.
In conclusion, to achieve efficient gene editing in porcine zygotes, CI of Cas9 protein with gRNA into zygotes after IVF has the possibility to be superior to that in oocytes before IVF. Furthermore, increasing the concentration of Cas9 protein and gRNA is effective in increasing mutation efficiency and generating biallelic mutant blastocysts. To reduce the mosaicism of microinjected zygotes, further optimization of the timing of CI and the concentration of CRISPR/Cas9 components is needed.
Acknowledgments
We thank the Nippon Food Packer, K. K. Shikoku (Tokushima, Japan), for supplying pig ovaries. This study was supported in part by the “Funds for the Development of Human Resources in Science and Technology” under MEXT, through the “Home for Innovative Researchers and Academic Knowledge Users (HIRAKU)” consortium, JSPS KAKENHI Grant Numbers JP17H03938, JP17K19325 and JP18K12062, and as a research project funded by SATREPS (Science and Technology Research Partnership for Sustainable Development).
References
- 1.Hammer RE, Pursel VG, Rexroad CE, Jr, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL. Production of transgenic rabbits, sheep and pigs by microinjection. Nature 1985; 315: 680–683. [DOI] [PubMed] [Google Scholar]
- 2.Galli C, Perota A, Brunetti D, Lagutina I, Lazzari G, Lucchini F. Genetic engineering including superseding microinjection: new ways to make GM pigs. Xenotransplantation 2010; 17: 397–410. [DOI] [PubMed] [Google Scholar]
- 3.Tan W, Proudfoot C, Lillico SG, Whitelaw CB. Gene targeting, genome editing: from Dolly to editors. Transgenic Res 2016; 25: 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laible G, Wei J, Wagner S. Improving livestock for agriculture - technological progress from random transgenesis to precision genome editing heralds a new era. Biotechnol J 2015; 10: 109–120. [DOI] [PubMed] [Google Scholar]
- 5.Keefer CL. Artificial cloning of domestic animals. Proc Natl Acad Sci USA 2015; 112: 8874–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014; 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, Whitelaw CB, Archibald AL. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog 2017; 13: e1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M, Koriyama M, Watanabe S, Ohtsuka M, Sakurai T, Inada E, Saitoh I, Nakamura S, Miyoshi K. Direct injection of CRISPR/Cas9-related mRNA into cytoplasm of parthenogenetically activated porcine oocytes causes frequent mosaicism for indel mutations. Int J Mol Sci 2015; 16: 17838–17856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato M, Kosuke M, Koriyama M, Inada E, Saitoh I, Ohtsuka M, Nakamura S, Sakurai T, Watanabe S, Miyoshi K. Timing of CRISPR/Cas9-related mRNA microinjection after activation as an important factor affecting genome editing efficiency in porcine oocytes. Theriogenology 2018; 108: 29–38. [DOI] [PubMed] [Google Scholar]
- 10.Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O’Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod 2014; 91: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan AW, Kukolj G, Skalka AM, Bremel RD. Timing of DNA integration, transgenic mosaicism, and pronuclear microinjection. Mol Reprod Dev 1999; 52: 406–413. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto M, Takemoto T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci Rep 2015; 5: 11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen TV, Tanihara F, Do L, Sato Y, Taniguchi M, Takagi M, Van Nguyen T, Otoi T. Chlorogenic acid supplementation during in vitro maturation improves maturation, fertilization and developmental competence of porcine oocytes. Reprod Domest Anim 2017; 52: 969–975. [DOI] [PubMed] [Google Scholar]
- 14.Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, Murphy CN, Carter DB, Hawley RJ, Prather RS. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 2002; 295: 1089–1092. [DOI] [PubMed] [Google Scholar]
- 15.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014; 42: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao L, Yang M, Wang X, Zhang Z, Wu Z, Tian J, An L, Wang S. Efficient biallelic mutation in porcine parthenotes using a CRISPR-Cas9 system. Biochem Biophys Res Commun 2016; 476: 225–229. [DOI] [PubMed] [Google Scholar]
- 17.Funahashi H, Stumpf TT, Cantley TC, Kim NH, Day BN. Pronuclear formation and intracellular glutathione content of in vitro-matured porcine oocytes following in vitro fertilisation and/or electrical activation. Zygote 1995; 3: 273–281. [DOI] [PubMed] [Google Scholar]
- 18.Kim BK, Cheon SH, Lee YJ, Choi SH, Cui XS, Kim NH. Pronucleus formation, DNA synthesis and metaphase entry in porcine oocytes following intracytoplasmic injection of murine spermatozoa. Zygote 2003; 11: 261–270. [DOI] [PubMed] [Google Scholar]
- 19.Laurincik J, Rath D, Niemann H. Differences in pronucleus formation and first cleavage following in vitro fertilization between pig oocytes matured in vivo and in vitro. J Reprod Fertil 1994; 102: 277–284. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014; 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 2015; 208: 44–53. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto M, Yamashita Y, Takemoto T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev Biol 2016; 418: 1–9. [DOI] [PubMed] [Google Scholar]
- 23.Vilarino M, Rashid ST, Suchy FP, McNabb BR, van der Meulen T, Fine EJ, Ahsan S, Mursaliyev N, Sebastiano V, Diab SS, Huising MO, Nakauchi H, Ross PJ. CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Sci Rep 2017; 7: 17472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheets TP, Park CH, Park KE, Powell A, Donovan DM, Telugu BP. Somatic cell nuclear transfer followed by CRIPSR/Cas9 microinjection results in highly efficient genome editing in cloned pigs. Int J Mol Sci 2016; 17: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Du Y, Shen B, Zhou X, Li J, Liu Y, Wang J, Zhou J, Hu B, Kang N, Gao J, Yu L, Huang X, Wei H. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep 2015; 5: 8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu HH, Zhao H, Qing YB, Pan WR, Jia BY, Zhao HY, Huang XX, Wei HJ. Porcine zygote injection with Cas9/sgRNA results in DMD-modified pig with muscle dystrophy. Int J Mol Sci 2016; 17: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 2014; 24: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park KE, Powell A, Sandmaier SE, Kim CM, Mileham A, Donovan DM, Telugu BP. Targeted gene knock-in by CRISPR/Cas ribonucleoproteins in porcine zygotes. Sci Rep 2017; 7: 42458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oji A, Noda T, Fujihara Y, Miyata H, Kim YJ, Muto M, Nozawa K, Matsumura T, Isotani A, Ikawa M. CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci Rep 2016; 6: 31666. [DOI] [PMC free article] [PubMed] [Google Scholar]