Fig. 9.
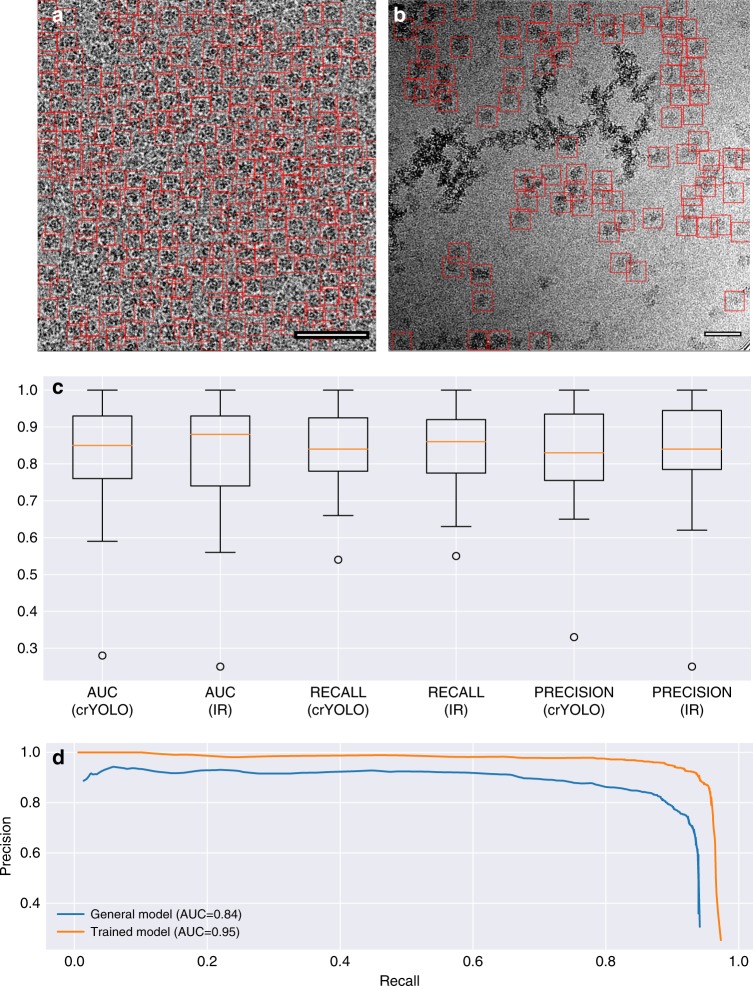
Generalized crYOLO network. a, b Particles selected on a representative micrograph of glutamate dehydrogenase (EMPIAR 10127) and RNA polymerase (EMPIAR 10190). None of the datasets were included in the set used for training the generalized crYOLO network. Scale bars, 50 nm. c AUC, recall, and precision of the datasets included into the general model evaluated for the crYOLO network architecture and the Inception-ResNet (IR) architecture (Supplementary Data 2). The box shows the lower and upper quartiles with a line as median. The whiskers represent the range of the data, whereas the points represent outliers. d Precision-recall curves for TcdA1 picked with either a network directly trained on the TcdA1 dataset (orange) and the general model but not on TcdA1 (blue) (Supplementary Data 3)