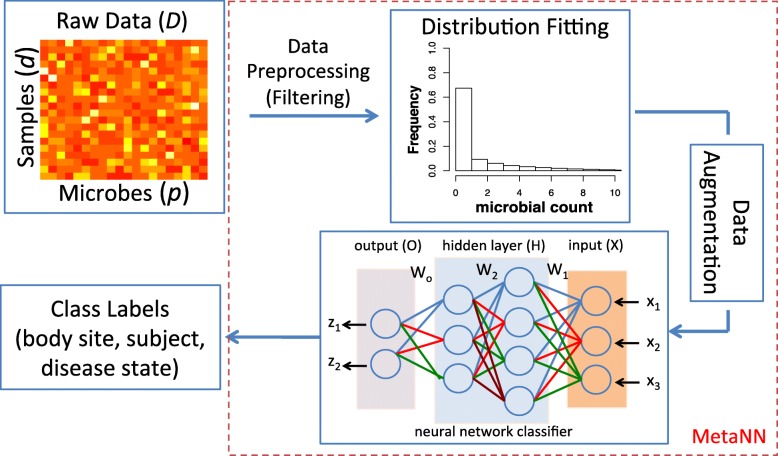
Fig. 1.
Our proposed MetaNN framework for the classification of metagenomic data. Given the raw metagenomic count data, we first filter out microbes that appear in less than 10% of total samples for each dataset. Next, we use negative binomial (NB) distribution to fit the training data, and then sample the fitted distribution to generate microbial samples to augment the training set. The augmented samples along with the training set are used to train a neural network classifier. In this example, the neural network takes counts of three microbes (x1,x2,x3) as input features and outputs the probability of two class labels (z1,z2). The intermediate layers are hidden layers each with four and three hidden units, respectively. The input for each layer is calculated by the output of the previous layer and multiplied by the weights (W1,W2,Wo) on the connected lines. Finally, we evaluate our proposed neural network classifier on synthetic and real datasets based on different metrics and compare outputs against several existing machine learning models (see Review of ML methods)