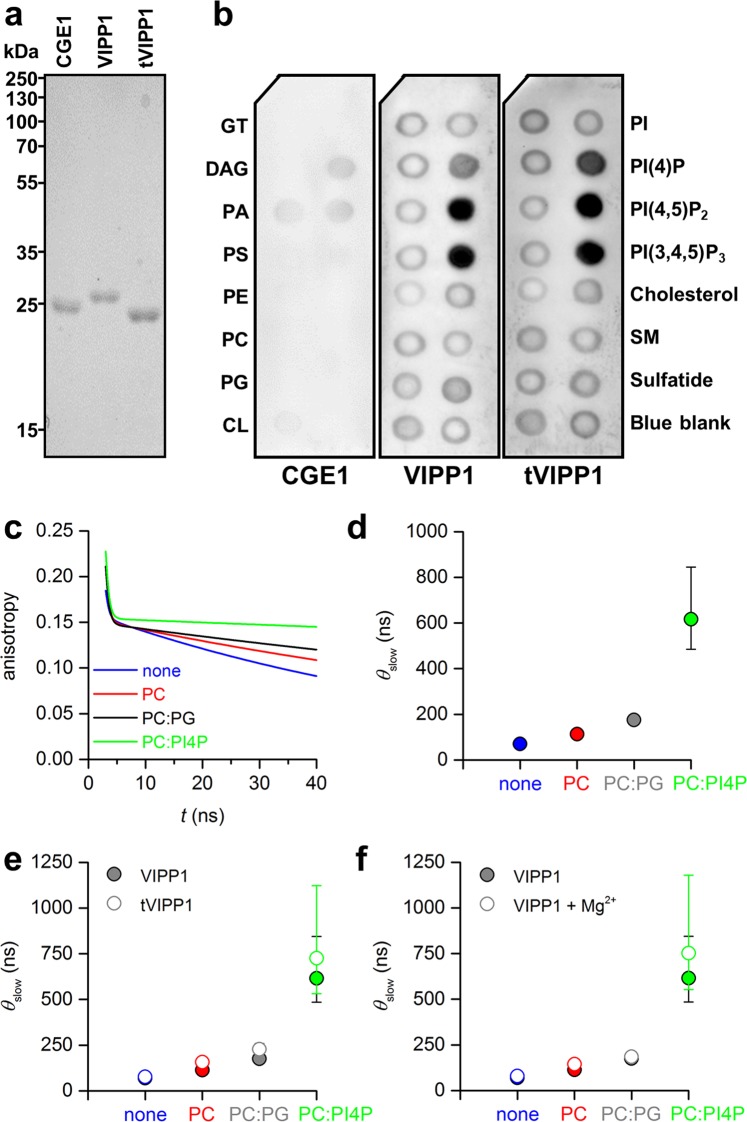
Figure 1.
The sizes of VIPP1-containing particles increase in the presence of PC:PI4P- liposomes. (a) Separation of 2 µg each of recombinant VIPP1, tVIPP1 and CGE1 on a 12% SDS-polyacrylamide gel, stained with Coomassie brilliant blue. (b) Protein–lipid overlay assay. Membranes spotted with different lipid species were incubated with the recombinant proteins followed by immunodetection of the bound proteins. Lipid species spotted were GT – glyceryl tripalmitate, DAG – diacylglycerol, PA – phosphatidic acid, PS – phosphatidylserine, PE – phosphatidylethanolamine, PC – phosphatidylcholine, PG – phosphatidylglycerol, CL – cardiolipin, PI – phosphatidylinositol, PIP – phosphatidylinositol phosphate, SM – sphingomyelin. Blue blank (Xylene Cyanol FF) is the negative control. (c) Fitted fluorescence anisotropy decay curves for VIPP1 without liposomes (none) and mixtures of VIPP1 with liposomes composed of PC, PC:PG (95:5), and PC:PI4P (95:5). (d) Best-fit values and 68% confidence intervals for the slow correlation times, θslow, which reflect the rotational mobility of VIPP1. Note that in most cases error bars are too small to be seen. (e) Best-fit values and 68% confidence intervals for θslow for the measurement shown in d and a measurement with tVIPP1. (f) Best-fit values and 68% confidence intervals for θslow for the measurement shown in d and a subsequent measurement performed in the presence of 10 mM Mg2+.