Abstract
Mental illness is increasingly recognized as both a significant cost to society and a significant area of opportunity for biological breakthrough. As -omics and imaging technologies enable researchers to probe molecular and physiological underpinnings of multiple diseases, opportunities arise to explore the biological basis for behavioral health and disease. From individual investigators to large international consortia, researchers have generated rich data sets in the area of mental health, including genomic, transcriptomic, metabolomic, proteomic, clinical and imaging resources. General data repositories such as the Gene Expression Omnibus (GEO) and Database of Genotypes and Phenotypes (dbGaP) and mental health (MH)-specific initiatives, such as the Psychiatric Genomics Consortium, MH Research Network and PsychENCODE represent a wealth of information yet to be gleaned. At the same time, novel approaches to integrate and analyze data sets are enabling important discoveries in the area of mental and behavioral health. This review will discuss and catalog into an organizing framework the increasingly diverse set of MH data resources available, using schizophrenia as a focus area, and will describe novel and integrative approaches to molecular biomarker discovery that make use of mental health data.
Keywords: translational bioinformatics, mental health, open access, biomarker discovery
Introduction
In 2013, mental illness was highly prevalent and estimated as incurring the highest financial burden among medical conditions in the United States, with spending estimated at $201 billion [1]. In light of the considerable cost to individuals and society, mental illness represents a compelling opportunity for discovery and improved patient care. As our ability to untangle biological mechanisms of disease grows, so too does our ability to leverage our richer understanding for better diagnoses, interventions and outcomes. In many areas of medicine, biomarker discovery is causing a shift toward biomarker-based diagnoses that promise better targeted and thus more effective therapies. Precision medicine approaches incorporating molecular and imaging biomarkers into therapeutic decision-making are emerging. Publicly available ‘big data’ resources like TCGA (The Cancer Genome Atlas) are being used and reused in numerous ways, with thousands of downstream citations [2]. Despite the magnitude of opportunity, translation has been slower in mental health (MH) than in other areas of health care [3]. In this article, we first describe the motivation as well as some challenges for the use of biomarkers in MH. We address one major challenge—findability of relevant resources—by providing a catalog of relevant data resources for biomarker discovery in MH, and a framework for their organization. Finally, we give an overview of existing approaches to biomarker discovery using publicly available data.
An exploration of biomarker in MH is especially timely in light of recent announcements from the National Institute of Mental Health (NIMH) exhorting a renewed focus on causal models of disease [4, 5]. Over the past 7 years, NIMH awards have shifted away from clinical research and trials and toward mechanistic biological understanding, coinciding with the NIMH’s launch of Research Domain Criteria (RDoC) in 2011, a framework emphasizing research into mechanisms (rather than clinically observable signs and symptoms) of mental illness [6].
Despite ample motivation, data reuse in MH research remains sluggish even in the presence of available biological resources and an emphasis on data sharing [7]. One important obstacle is the surprising difficulty in identifying available resources, related at least in part to an absence of a systematic approach to cataloging available resources. We seek to propose and use a systematic approach to organizing relevant resources. We then catalog data sets pertinent to MH biomarkers to facilitate secondary use of these data for computational biomarker discovery.
In addition to MH-focused resources and general resources that include MH conditions, a number of rich resources exist for specific MH disorders. However, inclusion of resources for every MH condition would far exceed space limitations for a single review. Therefore, in addition to general MH resources that span multiple disorders, we extend resource cataloging to a single MH disorder, schizophrenia (SCZ) and focus our biomarker discovery literature review on that disorder. SCZ is selected because it is one of the most studied MH disorders, puts heavy burden on the community and co-authors have conducted both large data annotations and various analyses in this area. It is also a prime example of a diagnostic concept well-recognized to be problematic and in need of updating [8, 9]. Biological exploration, biomarker discovery and elucidation of underlying mechanisms are key to addressing this issue. We have also limited our catalog to resources that are publicly available. Private or proprietary data sets or tools that are neither intended nor accessible for secondary research by independent researchers are beyond the scope of this review.
The mind-biology problem: a challenge for another day
The mind-body problem—What is the relationship between the mind (feelings, thoughts, beliefs) and the physical realm (matter, atoms, neurons)?—is commonly recognized in philosophy [10]. We stipulate, for the purposes of this discussion, that psychology becomes neurobiology once a biological mechanism is understood. As we gain insights into the neural basis of normal and abnormal behavior, syndromes historically described in terms of mental constructs can be described in terms of biological constructs. While we do not seek to tackle the philosophical question of whether all mental constructs can be adequately described in biological terms, we do assert that understanding biological mechanisms in MH is valuable, is likely to expand and will benefit from integrative research connecting behavior to biomarkers.
RDoC: ‘Outcomes to Causes and Back’
MH disorders typically include a spectrum of symptoms that affect emotions, thoughts and behaviors [4]. Moreover, two people can be diagnosed with a single disorder such as SCZ, despite having no overlapping symptoms. The NIMH RDoC initiative is an attempt to ‘develop, for research purposes, new ways of classifying mental disorders based on dimensions of observable behavior and neurobiological measures’ [4, 11]. The goal is to generate categories stemming from basic behavioral neuroscience, rather than starting with a highly heterogeneous illness definition and then seeking its neurobiological underpinnings. To this end, RDoC makes no reference to current DSM-based (Diagnostic and Statistical Manual of Mental Disorders) classification but instead proposes an alternative organizing scheme for linking behavior to underlying mechanisms. The RDoC approach is directly relevant to MH biomarkers because it aims to identify specific elements, such as mutations, genes, molecules, cells, pathways, physiological measures or behaviors associated with specific mental constructs across different disorders [3, 11].
Data, like life, is not always FAIR (Findable, Accessible, Interoperable, Reusable)
One of the expected and desirable results of the NIMH funding shift has been the generation of large biological data sets relevant to MH that are expected to be shared and reused. Recognizing the urgent need to improve infrastructure around data discoverability and reuse in the big data era, a group of stakeholders from academia, industry, funding organizations and publishers came together to design and endorse a set of measurable principles to act as guidelines for best practices in data sharing [12]. The resulting framework is known as FAIR principles—Findable, Accessible, Interoperable, Reusable. FAIR principles put particular emphasis on enhancing the ability for computers to find and use existing data. Findable refers to whether a researcher who would want to use the data set in question is able to discover that the data exist. This requires clear, persistent and searchable metadata. Accessible refers to whether the data are available to be downloaded. Are they retrievable through a standard communications protocol that enables authentication and authorization? Interoperable considers whether appropriate data and metadata standards are used for knowledge representation. Reusable addresses whether the data and provenance are represented in sufficient detail, with clear guidelines for usage.
While some researchers express concerns about reuse of clinical data in particular [13], many in the scientific community see significant benefit to be gained by data sharing and reuse [14]. The National Institutes of Health (NIH) has launched a Data Commons initiative to establish a virtual environment to facilitate the use, interoperability and discoverability of shared digital objects used for research [15]. This review focuses on those resources, data sets and publications that adhere to the spirit of the FAIR principles [12].
Biomarkers in mental health: What, why and how?
What is a ‘biomarker’ anyway?
A biomarker traditionally is defined as ‘a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological response’ [16]. Biomarkers can be generally classified as (1) diagnostic or trait markers that indicate the presence of a disease, (2) prognostic markers that indicate the likely course of a disease or (3) theranostic markers that predict how an individual is likely to respond to a certain treatment [17, 18]. As yet, no clinically actionable biomarkers have been approved for use in MH [11]. However, there is increasing recognition of the biological underpinnings of MH, the importance of biomarker discovery and the significant opportunity that MH poses in this regard. To this end, substantial research efforts have been devoted to biomarker discovery, and a number of publications describe promising leads [19–24]. Importantly, many of these studies have made their data publicly available to varying degrees, enabling secondary research and innovative approaches to analysis, in some cases through novel, integrative methods that could not have been done with the original data alone.
Biomarker types
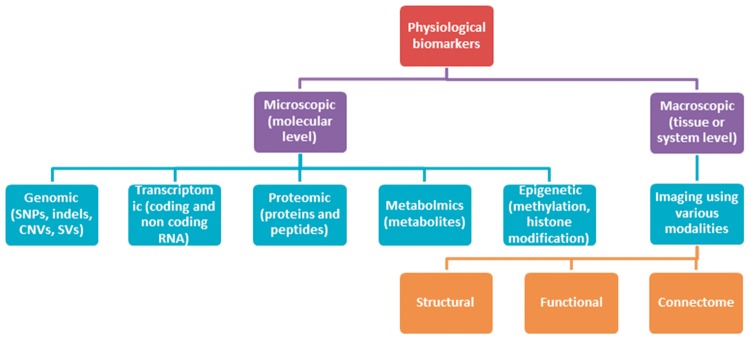
Physiological biomarkers span a wide range of modalities and data types and may be categorized as either microscopic or macroscopic in scale (Figure 1).
Figure 1.
Overview of micro- and macro-level biomarkers. indels, small insertions/deletions; SV, structural variants.
Micro-scale biomarkers: all things omics
Micro-scale biomarkers refer to biomarkers at the molecular level. The various and ever-increasing number of ‘-omic’ data-based biomarkers has been documented elsewhere [25, 26]. Genomic biomarkers generally refer to DNA sequence, including single-nucleotide variations (SNVs), copy number variations (CNVs), insertions, deletions, structural variants, etc. Transcriptomics refers to RNA expression, including both coding and noncoding RNA. Epigenomics refers to features of DNA other than the sequence itself, e.g. methylation, histone modification, etc. Proteomics refers to the presence, quantity and posttranslational modification state of proteins and peptides. Metabolomics refers to identification, quantification and ratios of various metabolites generated through the organism’s metabolism. Genomic and transcriptomic biomarkers have arguably received the most attention in the past decade, in part because they have become relatively low hanging fruit: microarrays and sequencing technologies make it fairly straightforward and increasingly inexpensive to make observations across the entire genome and transcriptome.
Macro-scale biomarkers: tissue and system level
Macro-scale biomarkers are observed at the tissue or system-level, generally through imaging technologies. Advances in brain imaging technology over past 20–30 years have enabled application to MH and illness. Commonly used imaging modalities include magnetic resonance imaging (MRI), magnetic resonance spectroscopy, positron-emission tomography (PET), single-photon emission computed tomography and diffusion tensor imaging (DTI). Other methods of neuroimaging involve recording of electrical currents or magnetic fields, for example electroencephalography (EEG) and magnetoencephalography (MEG). The additional biomarker types listed below are all macro-scale biomarkers.
Structural biomarkers of the brain
Structural imaging provides qualitative and quantitative information about the brain that describes the shape, size and integrity of gray and white matter structures in the brain. Typically, morphometric techniques measure the volume or shape of gray matter structures and white matter tracts. Structural MRI is used for identifying density or volume of brain matter, and DTI provides images of anatomical pathways and circuits especially of white matter [27].
Functional biomarkers of the brain
Whereas structural imaging provides static anatomical information, functional imaging provides dynamic physiological information [28]. Functional MRI (fMRI) and PET measure localized changes in cerebral blood flow related to neural activity, while EEG and MEG measure electrical currents and magnetic fields that vary with function.
The connectome: a ‘wiring diagram’ for the brain
The brain connectome defines the connectivity architecture and network organization of the neural components of the brain in terms of both structure and function. The connectome is represented as a large graph with nodes (brain regions) and edges (pathways) and has been enabled by advances in neuroimaging including structural MRI, fMRI and diffusion MRI [29]. Connectivity analysis based on graph theory is used to explore variations in the type and strength of connectivity between brain regions. Current evidence demonstrates alterations in both large-scale network and local network connectivity in mental health, and these alterations define distinct clinical and cognitive phenotypes [30].
Biomarker data resources
A framework for resource classification
A surprising challenge awaits a novice attempting integrative analyses: simply identifying what resources are available, how they relate to each other and what each one can and cannot provide is surprisingly difficult. In writing this review, we initially set out to catalog a list of publicly available data resources relevant to mental health. In the course of due diligence to identify these resources, certain categories and attributes emerged. Thus, our effort to catalog available resources also informed the creation of a candidate framework for classifying and organizing the different resource types.
Data resources can be classified as one (or sometimes more than one) of four high-level categories: (1) Organizational entity; (2) Initiative; (3) Platform; or (4) Data set (Figure 2). Examples of organizational entities include federal agencies, such as the NIMH, and nonprofit organizations, such as the Allen Institute for Brain Science [31]. Initiatives are activities or groups organized around activities aimed at creating, collecting or cataloging data for research. Examples include PsychENCODE, BioCADDIE and the Psychiatric Genomics Consortium (PGC) [32–34]. Data sharing platforms are Web-based applications that enable a researcher to search for data sets using metadata and to download the data. Examples include Sage Bionetworks’ Synapse platform or the Gene Expression Omnibus (GEO) [35, 36]. Finally, specific data sets may include data resulting from experimental assays, e.g. various data sets available in GEO, or curated knowledge bases like SZGR [28]. As shown in Figure 2, the relationships between different categories do not form a simple hierarchy but are instead are many-to-many. An initiative may be associated with one or more organizational entities, whether through funding or logistical or administrative support, while an organizational entity may be associated with one or more initiatives. A given organizational entity or initiative may rely on one or more platforms. A platform may contain (or point to) one or more data sets from one or more initiatives or organizational entities. A given data set is generally stored in one platform, but may also be accessed through other platforms, whether because it is replicated there or because some platforms serve as portals to federated data sets. These categories are not strictly mutually exclusive, and some resources blur the boundaries between them. For example, it can be hard to differentiate between an organization and an initiative. As a general rule, if an organization was created primarily for the purpose of creating or collecting data, we consider it an initiative. In addition, a curated knowledge base may import and redistribute some data sets on which it is based making it both a platform and a data set.
Figure 2.
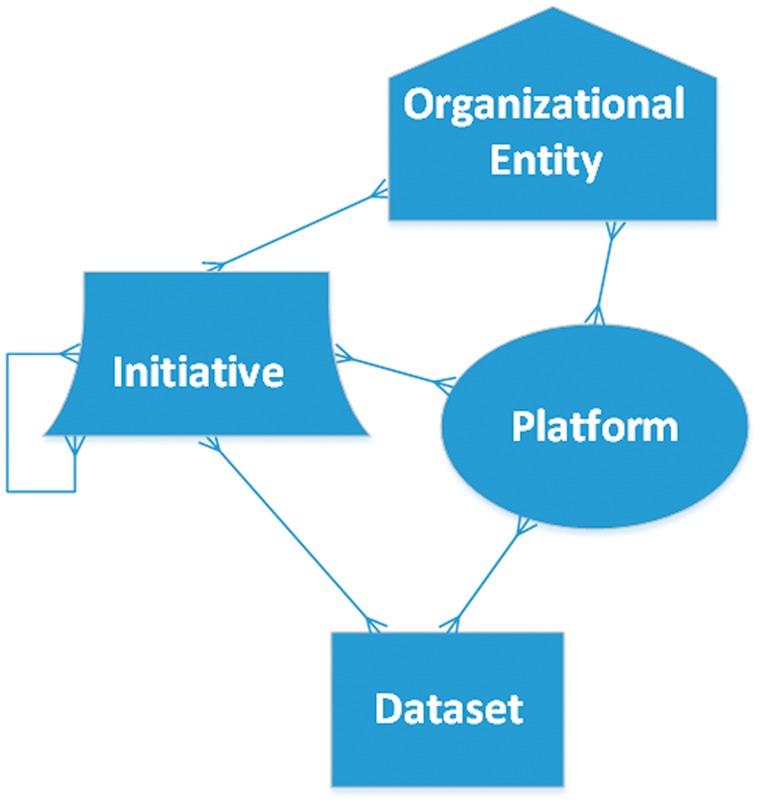
A framework for classification of data-related resources. Nodes denote resource types (Entities, Initiatives, Platforms and Data sets), and edges show the many-to-many relationships among them.
With respect to platforms, several attributes are especially salient. Some platforms such as Open fMRI focus on a single data type. Other platforms such as Synapse are meant to be general-purpose. In addition to storing different types of data, Synapse is disease-agnostic, storing data from many different diseases and medical domains. Other platforms, for example the Stanley Neuropathology Integrative Database focuses on a specific set of mental health conditions. Figure 3 shows where major data-sharing platforms relevant to mental health fall along the spectra of data-type specificity and disease focus. Some platforms, e.g. DataMed developed by the bioCADDIE project team for the NIH BD2K Data Discovery Index (DDI), are essentially portals to a federated collection of data sets that reside in still other platforms. Finally, some resources solely house data or information, while others are associated with biospecimens that may be available for generating additional data.
Figure 3.
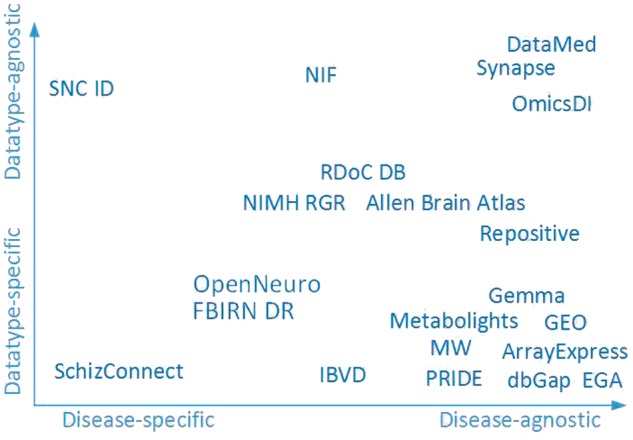
Visual representation of data platform attributes. See Table 1 for abbreviations.
Resource identification
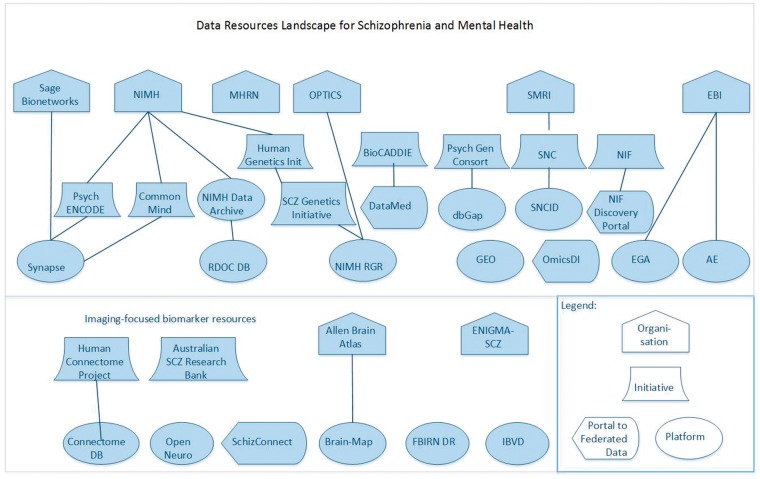
Because the topic of data for biomarker discovery in mental health is so broad, a simple PubMed query was not feasible. (For example, a query for ‘[mental health OR behavioral health OR psychiatric] AND [biomarker OR genomic OR imaging] AND data’ yields >25 000 hits.) A preliminary list of resources was established based on co-authors’ prior knowledge in the domains of open data, FAIR principles and MH. The list was then augmented through a series of searches in PubMed and Google Scholar using combinations and variations of the following terms: mental health, SCZ, open data, database, imaging, genomic, proteomic, metabolomic and biomarker. In addition to direct hits yielded by these terms, PubMed’s ‘similar articles’ provided valuable additional results. Finally, a search in BioCADDIE’s DataMed data search engine yielded additional sources. Inclusion criteria for resources were: (1) Scope includes one or more types of biomarker data (beyond clinical phenotype data); (2) Data accessibility, or at minimum some indication of how to request the data; and (3) Coverage of MH phenotypes, or in the case of disease-specific resources, SCZ. The resulting list of data resources and their metadata is provided in Table 1. Figure 4 gives a high-level landscape overview for the MH-specific organizational entities, initiatives and platforms and how they relate to each other.
Table 1.
Open data resources for biomarker discovery in mental health, particularly in schizophrenia
| Resource | Type | URL | Notes |
|---|---|---|---|
| Enhancing Neuro Imaging Genetics Through Meta Analysis (ENIGMA) | O | http://enigma.ini.usc.edu/ongoing/enigma-schizophrenia-working-group/ | The ENIGMA Network brings together researchers in imaging genomics to understand brain structure, function and disease, based on brain imaging and genetic data. Includes Schizophrenia Working Group (ENIGMA-SCZ) |
| NIMH | O | https://www.nimh.nih.gov/index.shtml | The institute within the NIH that focuses on mental health and disease. The NIMH is one of 27 institutes and centers within NIH, which is part of the US Department of Health and Human Services |
| Open Translational Science In Schizophrenia (OPTICS) | O | https://sites.google.com/site/opticsschizophrenia/home | A time-limited proof of concept pilot project designed to provide a forum for translational science based on Janssen clinical trial data made available to qualified investigators |
| Stanley Medical Research Institute (SMRI) | O | http://www.stanleyresearch.org/ | A nonprofit organization supporting research on the causes of, and treatments for, SCZ and bipolar disorder |
| Mental Health Research Network (MHRN) | O | http://hcsrn.org/mhrn | Consortium of 13 health system research centers dedicated to improving patient mental health through research, practice and policy. Supported by a cooperative agreement from the NIMH. The MHRN conducts pragmatic research in health systems serving over 12 million patients |
| Common Mind Consortium | I | http://commonmind.org | Public–private partnership to generate and analyze large-scale genomic data across several brain regions from human subjects with neuropsychiatric disease and to make these data and the associated analytical results broadly available to qualified investigators |
| Human Connectome Project (HCP) | I | http://www.humanconnectome.org/ | Large NIH-funded project for integrating genomics, behavior and brain imaging. Currently, high-resolution imaging data are available on 1200 individuals. Primary modalities measure brain activity (resting state fMRI and task-evoked fMRI), white matter integrity (diffusion imaging and T2 FLAIR) and oscillatory brain activity (EEG and) |
| NIMH Human Genetics Initiative | I | https://www.nimhgenetics.org/nimh_human_genetics_initiative/ | Intended to establish a national resource of clinical and diagnostic information and immortalized cell lines from individuals with SCZ, bipolar disorder or Alzheimer's disease and their relatives, available to qualified investigators for research on the genetic basis of these disorders |
| PsychENCODE | I | https://www.synapse.org//#! Synapse: syn4921369/wiki/235539 | Funded by the NIMH with the goal of accelerating discovery of noncoding functional genomic elements in the human brain and elucidating their role in the molecular pathophysiology of psychiatric disorders |
| Stanley Neuropathology Consortium (SNC) | I | http://www.stanleyresearch.org/brain-research/neuropathology-consortium/ | A collection of 60 brains, consisting of 15 each diagnosed with SCZ, bipolar disorder or major depression, and unaffected controls. Samples may be requested for research purposed. Associated data are available in the SNC Integrative Database (SNCID)—see below |
| Psychiatrics Genomics Consortium (PGC) | I | http://www.med.unc.edu/pgc | Founded in 2007, the PGC includes over 800 investigators from 38 countries with the goal of conducting meta- and mega-analyses of genomic data for psychiatric disorders. The initial focus was on autism, attention-deficit hyperactivity disorder, bipolar disorder, major depressive disorder and SCZ. More recently, the scope has expanded to other conditions and other types of genetic variation beyond SNVs |
| Neuroscience Information Framework (NIF) | I/P | https://neuinfo.org/ | An NIH-funded framework for identifying, locating, relating, accessing, integrating and analyzing information from the neuroscience research enterprise. NIF has come to refer to both this initiative and the set of tools and platforms that make up that framework including the registry of electronic resources and the discovery portal for searching those resources. NIF includes >4500 curated resources and access to > 100 databases |
| Allen Brain Atlas/Data Portal | I/P | http://human.brain-map.org/ | The Allen Institute for Brain Science is dedicated to understanding how the human brain works in health and disease. The Allen Human Brain Atlas integrates anatomic and genomic information across the brain. Data modalities include MRI, DTI, histology and gene expression data derived from both microarray and in situ hybridization (ISH) approaches. Microarray data are spatially mapped to the MRI. Complete microarray and RNA-seq data are available for six human brains. ISH data are available for ∼50 SCZ brains |
| NIMH Repository and Genomics Resource (RGR) | P | https://www.nimhgenetics.org/available_data/schizophrenia/ | Includes 100+ studies, including CommonMind, PsychENCODE. Formerly the Center for Collaborative Genomic Studies on Mental Disorders, the RGR was established in 1998 through the NIMH Human Genetics Initiative to leverage and increase the value of human genetic samples and data produced through NIMH-funded research. It contains a collection of > 150 000 well-characterized, high-quality patient and control samples from patients with a range of mental disorders. The RGR’s Biologic Core and a Data Management Core are external to NIH |
| Function Biomedical Informatics Research Network Data Repository (FBIRN DR) | P | fbirnbdr.nbirn.net: 8080 (BROKEN) | FBIRN was initially focused on assessing major sources of variation of fMRI data generated across different scanners. The FBIRN Phase 1 data set consists of a traveling subject study of five healthy subjects, each scanned on 10 different 1.5 to 4 T scanners. The FBIRN Phase 2 and Phase 3 data sets consist of subjects with SCZ or schizoaffective disorder along with healthy comparison subjects scanned at multiple sites. The BIRN Data Repository (BDR) includes imaging, clinical, cognitive and physiological data |
| OpenNeuro (previously OpenfMRI) | P | https://openneuro.org/(https://openfmri.org/) | A neuroimaging repository to enable reproducible analysis and data sharing. Started in 2010, it initially focused only on task-based MRI, but is now open to all forms of neuroimaging data, reflected in the name transition from OpenfMRI to OpenNeuro. Data are anonymized before distribution to protect the confidentiality of participants and distributed using a Public Domain license |
| Research Domain Criteria Database (RDoC DB) | P | https://data-archive.nimh.nih.gov/rdocdb/ | A data repository for the harmonization and sharing of research data related to the RDoC initiative and mental health research more generally. The actual platform uses software designed to host the NIH’s National Database for Autism Research (NDAR) |
| SchizConnect | P | http://schizconnect.org/ | Federated access to several neuroimaging databases with images acquired on SCZ subjects. Data sources include FBIRN, NUSDAST, COINS and MCIC (maintained by the Mental Illness and Neuroscience Discovery Institute, now the Mind Research Network). More than 1100 subjects with >1000 have imaging data, including resting state fMRI, task-related fMRI, structural and diffusion imaging |
| SNCID | P | http://sncid.stanleyresearch.org/ | Web-based tool for exploring neuropathological traits, gene expression and associated biological processes in psychiatric disorders generated by the SNC within the SMRI |
| Australian Schizophrenia Research Bank | P | http://www.schizophreniaresearch.org.au/bank/ | A research database and storage facility that links clinical and neuropsychological information, blood samples and structural and fMRI brain scans from people with SCZ and healthy nonpsychiatric controls, and currently has data on ∼900 cases and 900 controls |
| Internet Brain Volume Database (IBVD) | P | http://ibvd.virtualbrain.org/ | Centered around publications as the central data structure, IBVD is a Web-based searchable database of brain neuroanatomic volumetric observations that enables electronic access to the results in the published literature |
| dbGap | P | https://www.ncbi.nlm.nih.gov/gap | Developed by the NIH’s NCBI to archive and distribute the data and results from studies that have investigated the interaction of genotype and phenotype. While the focus is on genomic data, other data types are included as well, for example metabolomic data and laboratory values |
| Metabolights | P | http://www.ebi.ac.uk/metabolights/ | A database for Metabolomics experiments and derived information. Metabolights is the slightly more established European counterpart to the NIH’s MW and the recommended metabolomics repository for a number of top journals |
| DataMed | P | http://datamed.org/ | Data search engine portal to enable users to search for data across different repositories developed for the NIH BD2K DDI by the bioCADDIE project team. The initial prototype release (v2.0) features a set of data repositories selected by the bioCADDIE team, with a form to suggest additional repositories for inclusion |
| Metabolomics Workbench (MW) | P | http://www.metabolomicsworkbench.org/ | A repository for metabolomics data and metadata, MW provides analysis tools and access to metabolite standards, protocols, tutorials and training |
| PRIDE | P | https://www.ebi.ac.uk/pride/archive/ | A centralized, standards compliant, public data repository for proteomics data, including protein and peptide identifications, posttranslational modifications and supporting spectral evidence. Most of the data sets related to mental health disorders in PRIDE are derived from animal models |
| Synapse | P | https://www.synapse.org/ | Sage Bionetworks’ software platform for data sharing and provenance tracking. Synapse enables researchers to carry out, track and communicate research in real time and enables co-location of scientific content (data, code, results) and narrative descriptions of that work. The platform is agnostic regarding biomedical domain or data type and hosts a number of different file types and projects funded by a number of different sources |
| GEO | P | https://www.ncbi.nlm.nih.gov/geo/ | An international public repository developed by the NIH NCBI that archives and freely distributes microarray, next-generation sequencing and other high-throughput functional genomics data submitted by the research community |
| AE | P | https://www.ebi.ac.uk/arrayexpress/ | The European counterpart to GEO. AE is an archive of functional genomics data from high-throughput functional genomics experiments. A subset of experiments is imported from GEO, while others are submitted directly |
| GEMMA | P | http://www.chibi.ubc.ca/Gemma/ | Gemma is a website, database and a set of tools for the meta-analysis, re-use and sharing of genomics data, currently primarily targeted at the analysis of gene expression profiles |
| OmicsDI | P | http://www.omicsdi.org | Enables data set discovery across omics data resources spanning eight international repositories, including both open and controlled access data resources. The resource provides key metadata for each data set and uses this metadata to enable search capabilities and identification of related data sets. OmicsDI helps researchers to idenitfy groups of related, multi-omics data sets across repositories |
Note: Type: O, organizational entity; I, initiative; P, platform.
Figure 4.
Overview of landscape of organizational entities, initiatives and data sharing platforms.
A number of potentially useful resources were deemed out of scope for this review because they lacked either -omics data (e.g. National Database for Clinical Trials Related to Mental Illness, NDCT, Yale Open Data Access Project YODA [37]), or psychiatric phenotype data (Exome Aggregation Consortium, ExAC [38], Genotype-Tissue Expression (GTEx) project [39]).
Data sets
Genomic data
The two main data repositories for gene expression or transcriptomic data are GEO and ArrayExpress (AE). GEO is an international public repository developed by the US NIH’s National Center for Biotechnology Informatics (NCBI) that archives and freely distributes microarray, next-generation sequencing and other high-throughput functional genomics data submitted by the research community [35]. AE is the Europe-based repository, hosted by the European Bioinformatics Institute within the European Molecular Biology Laboratory (EMBL-EBI). Data are imported from GEO into AE on a weekly basis making GEO a subset of AE. To be uploaded to these data repositories, data sets need to be in a specific format, such as GEOarchive, SOFT or MINiM. They must also include appropriate metadata about the clinical and experimental data. Both AE and GEO enable programmatic access to data via tools like R/Bioconductor. Data sets in GEO therefore are able to satisfy the F (findable) and R (reusable) FAIR criteria. Gene expression data in GEO are generally considered to be de-identified, and are thus freely available for public use.
There has been an increase in genomic profiling of data related to MH in the last few years. Taking SCZ as an example, a search in AE for published SCZ data sets shows 92 data sets in humans (Supplementary Table S1). Only two data sets were published in 2007 as compared with 11 data sets published in 2016. Until 2010, the majority of published data sets concerned transcriptome profiling. In 2012 and 2013, other genomic methods had gained popularity including methylation and next-generation sequencing technologies, exploration of noncoding regions and gene expression and splicing. Since 2014, many studies have been published using newer genomic platforms including chromatin immunoprecipitation sequencing, RNA sequencing (RNA-seq) and microRNA-seq, amounting to an approximate 15 published studies in 2014, 13 studies in 2015 and 11 studies in 2016. As of July 2017, we found that of the 92 data sets, 80 had been cited in one or more subsequent publications. Using Google Scholar queries on data set accession identifiers, it was determined that these 80 publications have been cited 6710 times (Supplementary Table S1). Note that citation does not necessarily imply analysis: many publications called attention to the existence of a data set without performing any additional analysis.
NCBI’s database of Genotypes and Phenotypes (dbGaP) contains archived data and results from studies that have investigated the association between genotype and phenotype in Humans [40]. The European equivalent is the EGA (European Genome-phenome Archive) [41]. As with the gene expression repositories, data sets need to be in a specific format along with minimal metadata to be submitted into dbGaP or EGA. Note that these repositories contain sequencing data that are unique to the individuals from whom they were derived, and thus cannot be considered completely de-identified. Users must therefore submit a data request form detailing the goals of their project and how they intend to use the data and observe data use policy for approval by a data request committee. This approach has implications for meeting the accessibility aspect of FAIR criteria but represents a balance between data accessibility and data privacy for research participants. dbGaP contains a number of MH-related data sets, including SCZ. Of the 154 studies returned based on a query for the term ‘schizophrenia’, only a small subset was targeted at SCZ as determined by manual inspection. In this case, findability is hampered by the number of false positives (Table 2). The vast majority of the 24 studies returned in a search for ‘schizophrenia’ in EGA are either focused on SCZ or have some number of samples included with a SCZ diagnosis.
Table 2.
SCZ data sets in dbGaP
| Data set ID | Name | # Participants | Platform | Publication (PMIDs) | Citations | Data type |
|---|---|---|---|---|---|---|
| phs000979.v1.p1 (PRJNA293910) | Gene Expression in Postmortem DLPFC and Hippocampus from Schizophrenia and Mood Disorders | 914 | HumanHap650Yv3.0, Human1M-Duov3_B, Human HT-12 Expression Bead Ch | 28070120 | [4] | SNP array, mRNA expression |
| phs000473.v2.p2 (PRJNA157243, PRJNA94281) | Sweden-Schizophrenia Population-Based Case-Control Exome Sequencing | 12 380 | SureSelect Human All Exon v.1 Kit, SureSelect Human All Exon v. | 22641211 | [15] | WES |
| phs000738.v1.p1 | Exome Sequencing in Schizophrenia Families | 216 | SeqCap EZ Human Exome Library v2.0 | 23911319, 24317315 | [1] | WES |
| phs000687.v1.p1 | Bulgarian Schizophrenia Trio Sequencing Study | 1826 | SureSelect Human All Exon v.2 Kit, SureSelect Human All Exon v3-50Mb, SeqCap EZ Human Exome Library v2.0 | 23040492, 22083728, 24463507 | [1] | WES, SNP Genotype |
| phs000608.v1.p1 | Whole-Genome Profiling to Detect Schizophrenia Methylation Markers | 1459 | MBD-seq | 23244307 | [42] | Methylation |
| phs000448.v1.p1 | Genetics of Schizophrenia in an Ashkenazi Jewish Case-Control Cohort | 3096 | HumanOmni1-Quad_v1-0_B | [4] | SNP array | |
| phs000021.v3.p2 | Genome-Wide Association Study of Schizophrenia | 5064 | AFFY_6.0 | 16400611 | [43] | SNP array |
| phs000167.v1.p1 | Molecular Genetics of Schizophrenia-nonGAIN Sample (MGS nonGAIN) | 3029 | AFFY_6.0 | 16400611 | [44] | SNP array |
Proteomic and metabolomic data sets
EBI’s Metabolights and NCBI’s Metabolomics Workbench (MW) are two major metabolomics data repositories. Metabolights has no SCZ data sets; MW has one but data are not downloadable. A limited number of data sets appear to be available for other mental health phenotypes such as Alzheimer’s disease and autism spectrum disorder. PRIDE (PRoteomics IDEntifications), the leading proteomics data repository, has only three SCZ-related data sets: two in rat models and one in a mouse model. Data sets related to other MH disorders are similarly limited and largely generated from animal models.
Imaging repositories
SchizConnect is a federated portal that integrates data from three neuroimaging consortia on SCZ: FBIRN's Human Imaging Database (HID), MRN's Collaborative Imaging and Neuroinformatics System (COINS) and the Northwestern University Schizophrenia Data and Software Tool (NUSDAST) project [45].
A number of general purpose brain imaging repositories exist, such as OpenNeuro (formerly OpenfMRI) [46], and the Neuroimaging Informatics Tools and Resources Clearinghouse Image Repository (NITRC IR) [47]. However, to date, the publicly available data sets in those resources appear to be more cognition-oriented (e.g. classification learning, visual and auditory functions, attention) than psychiatric. Notable exceptions in SCZ include [42, 48].
The Functional Connectomes Project is also available through NITRC [49]. It comprises data from >1400 healthy subjects who underwent fMRI scans that assessed their brain activity when their minds were at rest. Included in the 1400 is a subset known as the COBRE (Center for Biomedical Research Excellence) data set, which includes anatomical and functional MR data from 72 patients with SCZ and 75 healthy controls. These data have been analyzed in a number of different ways by different groups [50–53].
Curated knowledge bases
SZGR [28], SZGene [44] and SZDB [54] are three distinct but significantly overlapping knowledge repositories that include curated information regarding SCZ-related genes and data sets. The SZGR, available since 2009, is a ‘one-stop shop’ for genes and variants in SCZ, along with their function, regulation and drug information. It was created through systematic review and curation of multiple lines of evidence and includes ∼4200 common mutations and ∼1000 de novo mutations [28, 55]. SZGene is affiliated with the Schizophrenia Research Forum and contains data from 1700 studies. It enables the user to search by gene, protein, polymorphism, study or keyword to return the specific publications addressing those features [44]. However, the resource only contains data from studies before 2012 and is no longer supported. (Unfortunately, this is not uncommon for resources that require maintenance over time.) Finally, SZDB includes genomic, transcriptomic, molecular network data and functional annotations [54].
The DisGeNET database (http://www.disgenet.org) integrates human gene–disease associations from various expert curated databases and text-mining-derived associations including Mendelian, complex and environmental diseases. A search in July 2017 for genes and single-nucleotide polymorphisms (SNPs) associated with SCZ yielded 1871 and 1635, respectively.
Genome-wide association studies and beyond: innovative, integrative approaches to biomarker discovery in mental health
At the most basic level, data sharing can enable integrative analysis for biomarker discovery by allowing researchers to combine two or more comparable data sets to increase statistical power through increased sample size. Researchers have developed many creative methods to combine data, enabling the discovery of patterns not apparent when analyzing just a single data type. Genotype data, including those identified through both microarrays and next-generation sequencing, can be combined with other data types such as protein–protein interaction networks, biological pathways, gene expression and co-expression, methylation and microRNA regulation data [43, 56–60]. In some cases, researchers have been able to combine three or more data types in creative ways for biomarker discovery [61, 62].
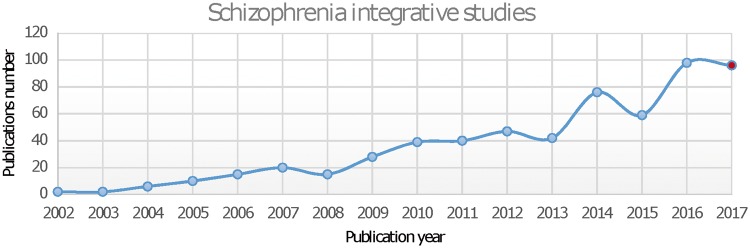
Most studies start with the purpose of discovering novel variants and then make use of public resources to validate and support their initial discovery. Methods may be categorized at the gene level [63–66], pathway level [67] and network level [68–70]. Another way to categorize the methods is based on the multi-omics data. Some integrative studies involve only genetics and eQTL (expression quantitative trait loci) data [71], while others are more comprehensive, involving multiple genome-wide association studies (GWAS) and/or other dimensional data [72, 73]. Recently, with the dramatic increase of GWAS data, especially by the PGC, an increasing number of studies have been published for integrative analyses using multiple-disorders or multiple-omics data, aiming to identify shared or unique genetic variants among different MH disorders [74]. To generate an overview of these integrative studies, we used PubMed to systematically search for integrative studies using keywords listed in Table 3. In total, we obtained 595 publications for integrative studies of SCZ. A majority of them (497) were published after 2010, likely due in part to the curation of omics data in recent years. As shown in Figure 5, the publication of integrative studies in SCZ has been increasing sharply in recent years, mostly in the category of eQTL. Recurring themes among these integrative methods include overlap between variants from different omics modalities and randomization and permutation tests for statistical significance.
Table 3.
Keywords and counts for integrative biomarker studies in schizophrenia published before May 2017
| Keywords | N |
|---|---|
| schizophrenia [TIAB] AND GWAS AND expression | 285 |
| schizophrenia [TIAB] AND SNP AND expression | 242 |
| schizophrenia [TIAB] AND GWAS AND network | 140 |
| schizophrenia [TIAB] AND SNP AND network | 75 |
| schizophrenia [TIAB] AND GWAS AND methylation | 36 |
| schizophrenia [TIAB] AND GWAS AND eQTL | 35 |
| schizophrenia [TIAB] AND SNP AND integrative | 32 |
| schizophrenia [TIAB] AND GWAS AND quantitative traits | 26 |
| schizophrenia [TIAB] AND GWAS AND transcriptome | 26 |
| schizophrenia [TIAB] AND SNP AND methylation | 20 |
| schizophrenia [TIAB] AND SNP AND eQTL | 19 |
| schizophrenia [TIAB] AND SNP AND quantitative traits | 11 |
| schizophrenia [TIAB] AND SNP AND transcriptome | 10 |
| schizophrenia [TIAB] AND GWAS AND integrative | 5 |
| schizophrenia [TIAB] AND SNP AND transcriptome | 4 |
| schizophrenia [TIAB] AND genotyping AND transcriptome | 3 |
| schizophrenia [TIAB] AND SNP AND ATAC-seq | 1 |
Figure 5.
Publication summary of SCZ integrative studies.
Note: Publications in 2017 were estimated based on the data between January and May in 2017.
Big big-data: large-scaleGWASs
Since 2008, GWAS have reported a number of genetic variants associated with SCZ [63–66]. The Schizophrenia Working Group of the PGC conducted the largest GWAS in SCZ to date (36 989 SCZ and 113 075 controls) and identified 108 loci [75]. The largest ancestrally and phenotypically homogeneous GWAS study of SCZ (11 260 cases and 24 542 controls) reported 50 novel SCZ risk loci [76].
Despite these large-scale studies, the genes or functional DNA elements through which these variants exert their effects remain unknown. An emerging trend is to integrate multidimensional data from genetics, epigenetics and transcriptomics to prioritize biomarkers and also to understand the underlying mechanisms by which they act.
Combining expression and sequence data: eQTL
Several large-scale studies have markedly expanded the scope of known eQTLs [77–79]. One recent example integrates SCZ postmortem brain gene expression with GWAS signals at the pathway levels [80]. Building on these eQTL resources as well as gene expression profiles, candidate genes have been identified by integrative studies [81–85]. Using expression data from 647 postmortem human brain samples collected by Stanley Medical Research Institute and the GTEx project, the gene complement component 4 (C4) in major histocompatibility complex region was identified as contributing to SCZ risk [86]. In addition, the CommonMind Consortium (CMC) generated RNA-seq data from postmortem dorsolateral prefrontal cortices from 258 subjects with SCZ and 279 controls, and they identified a list of genes whose expression was significantly affected by SCZ risk variations [87]. Using the CMC data, a list of brain splicing quantitative trait loci was identified that are causally associated with SCZ [88]. In another study, SCZ risk genes were identified using summary data from GWAS and eQTL in which gene expression data were generated from 5311 peripheral blood samples [89]. More examples include valuable eQTLs located at NMDAR [90], CTCF and CACNB2 [91], and 17q25 locus [92], most of which used public eQTL data such as those from GTEx. These eQTLs are mostly common variants. Rare noncoding SCZ risk variants were also identified [93].
Combining genomic data with multiple phenotypes: pleiotropy
Genomic data combined with multiple phenotypes can enable the discovery of pleiotropy-associated genes, i.e. alleles that impact two or more apparently unrelated effects. The PGC provides a good example of this. They initially, and intentionally, focused studies on five major psychiatric disorders: autism, attention-deficit hyperactivity disorder, bipolar disorder, major depressive disorder and SCZ [74, 94]. A number of clinical features transcend these disease classifications, and previous research had suggested overlap in familial and genetic liability for different combinations of these disorders [74].
Combining genomic and imaging data
Another common integrative approach in the study of MH is the combination of genomic data with imaging data. The Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) consortium provides tools and protocols to meta-analyze genome-wide and neuroimaging data from research teams worldwide [95]. The consortium does not require participating investigators to contribute raw data nor provide access to such data for research or public use. Instead, ENIGMA provides standardized protocols with predefined covariates to allow sites to conduct GWAS and imaging studies locally and report meta-analyzed data, which are made publicly available through an interactive visualization tool, ENIGMA-Vis [96]. For example, the ENIGMA SCZ working group conducted a collaborative, prospective meta-analysis of neuroimaging data from >4500 study participants (2028 were subjects with SCZ and 2540 were healthy control) across 15 sites [97].
Combining genomic and epigenetic data
DNA methylation is important for epigenetic regulation of gene expression. The 108 SCZ-GWAS risk loci identified by the PGC [75] were evaluated systematically as methylation quantitative trait loci in postmortem prefrontal cortex from 191 SCZ and 335 controls [98], 689 SCZ and 645 controls [99] and 1163 postmortem brains of European ancestry [100]. The loci were also systematically analyzed using 166 human fetal brains [101]. The evidence showed a much stronger differential DNA methylation enrichment in genes associated with SCZ, even using a medium sample size (<100) of postmortem brains [102].
Transposase Accessible Chromatin followed by sequencing (ATAC-seq) is another promising technique that can be used to map chromatin accessibility. Recently, ATAC-seq has been used to study spatiotemporal regulation of gene expression of neuronal and nonneuronal nuclei isolated from frozen postmortem human brain to map chromatin accessibility for SCZ risk loci [103].
Concluding remarks
Our review of data resources and integrative biomarker discovery in MH with a focus on SCZ suggests a recent increase in the number and quality of resources and an even more recent growth in their use. Indeed, we are starting to see high-profile papers that leverage some of these existing data sets. For example, a recent paper in Nature Genetics described a GWAS study in 36 000 individuals of Chinese ancestry that was combined with data from the PGC [75] to perform trans-ancestry meta-analyses yielding 30 novel risk loci for SCZ [104]. However, there remains significant untapped potential: many resources that could be made available under FAIR principles are not, and those resources that are available remainunderused. While we leave other MH disorders for a future paper, we do not believe the results will be significantly different for anxiety or depression, or for most other disorders in mental health with the possible exception of autism spectrum disorder, where a concerted national effort by both public and private entities has created a large concentration of findable and accessible shared data [105, 106].
The challenges facing those seeking to reuse resources for integrative research are numerous and formidable, but we believe that many seeking to enter the fray are tripping over the threshold of findability. We hope that both the suggested organizing framework and the catalog of resources presented here will help new entrants into the space cross the threshold successfully and focus on more substantive challenges of data integration and analysis. We also believe the proposed framework is useful more broadly in other biomedical domains to facilitate categorization and dissemination of information about data resources to support emerging precision medicine initiatives.
As noted recently in a memo from Dr Joshua Gordon, director of NIMH, understanding the underlying biology of MH is more important than ever, and increasingly within reach given recent technological developments [4]. A consensus on FAIR principles, the NIH push toward data sharing, and NLM support for best practices, mean those who continue to develop innovative approaches to the vast and ever-increasing amount of publicly available data will help the rest of us gather valuable insights about mental health diagnosis and treatment.
Supplementary Material
Acknowledgements
The authors wish to thank Dr Piper Ranallo for her passion and perseverance in catalyzing creation of the AMIA Mental Health Informatics Working Group out of which this article emerged and Jyotishman Pathak for valuable comments on early drafts of the manuscript. The authors also thank the anonymous reviewers for their insightful comments and helpful suggestions to strengthen this review.
Jessica D. Tenenbaum is an assistant professor of Translational Biomedical Informatics in the Department of Biostatistics and Bioinformatics at the Duke University School of Medicine. She is co-founder and chair of the American Medical Informatics Association (AMIA)’s Mental Health Informatics Working Group.
Krithika Bhuvaneshwar is a research associate and project manager at the Innovation Center for Biomedical Informatics (ICBI) at Georgetown University. She has expertise in bioinformatics analysis and systems biology research, and combines her interdisciplinary skills in bioinformatics and biostatistics for numerous projects.
Jane Gagliardi is an associate professor of psychiatry and behavioral sciences and an associate professor of Medicine at Duke University School of Medicine and serves as vice chair for education and residency training director in Psychiatry. Her main areas of interest and expertise are clinical psychiatry, clinical medicine, patient safety and quality and the impact of electronic technology on patient care and education.
Kate Fultz Hollis is a research associate and instructor in the Department of Biomedical Informatics and Clinical Epidemiology at Oregon Health and Science University. She specializes in clinical research informatics and medical research data access.
Peilin Jia is an assistant professor in Center for Precision Health, School of Biomedical Informatics, The University of Texas Health Science Center at Houston. She co-directs the Bioinformatics and Systems Medicine Laboratory.
Liang Ma is a bioinformatics postdoctoral fellow in the Bioinformatics and Systems Medicine Laboratory (BSML), Center for Precision Health, School of Biomedical Informatics, the University of Texas Health Science Center at Houston.
Radhakrishnan Nagarajan is an associate professor of biomedical informatics in the College of Medicine, University of Kentucky. His research involves developing novel analytics for knowledge discovery from heterogeneous molecular and health-care data.
Gopalkumar Rakesh is a physician-scientist in training with the Department of Psychiatry at Duke University Medical Center. His main areas of interest and expertise are clinical psychiatry, brain stimulation and big data analytics.
Vignesh Subbian is an assistant professor in the Department of Biomedical Engineering and the Department of Systems and Industrial Engineering at the University of Arizona.
Shyam Visweswaran is an associate professor of biomedical informatics and the Intelligent Systems Program at the University of Pittsburgh. He is the director of Clinical Informatics for the Department of Biomedical Informatics, the director of the Center for Clinical Research Informatics and the director of the Biomedical Informatics Core of the University of Pittsburgh Clinical and Translational Science Institute.
Zhongming Zhao is the chair and professor for Precision Health and director of Center for Precision Health, School of Biomedical Informatics, the University of Texas Health Science Center at Houston. He directs the Bioinformatics and Systems Medicine Laboratory.
Leon Rozenblit is the Founder and CEO of Prometheus Research, LLC, a research informatics services and technology company with a concentration in mental health informatics. He served as the informatics lead on a number of large-scale mental health research initiatives including the Simons Simplex Collection and the Autism Biomarker Consortium for Clinical Trials.
Key Points
The growing number of data sets available to researchers in mental and behavioral health enables secondary analysis and novel integrative methods for biomarker discovery.
We propose a framework for organizing and classifying publicly available resources for biomarker discovery in mental health using SCZ as an example.
Many potential resources are not yet compliant with FAIR data-sharing principles, and currently available resources remain underused.
While no clinically actionable biomarkers have yet been identified, a confluence of policies, initiatives and technological advances puts us at a potential inflection point for accelerating discovery and advancement in the field of MH.
Funding
The National Institutes of Health (grant numbers UL1TR001117 to J.D.T., R01LM012095 to S.V. and R01LM012806 to P.J. and Z.Z.).
References
- 1. Roehrig C. Mental disorders top the list of the most costly conditions in the United States: $201 billion. Health Aff 2016;35(6):1130–5. [DOI] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network, Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455(7216):1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kapur S, Phillips AG, Insel TR.. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol Psychiatry 2012;17(12):1174.. [DOI] [PubMed] [Google Scholar]
- 4. Gordon J. RDoC: Outcomes to Causes and Back Bethesda, MD: NIH, 2017. [Google Scholar]
- 5. Reardon S. US mental-health agency’s push for basic research has slashed support for clinical trials. Nature 2017;546:339. [DOI] [PubMed] [Google Scholar]
- 6. Insel T, Cuthbert B, Garvey M, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry 2010;167(7):748–51. 10.1176/appi.ajp.2010.09091379 [DOI] [PubMed] [Google Scholar]
- 7. Johnson EC, Border R, Melroy-Greif WE, et al. No evidence that schizophrenia candidate genes are more associated with schizophrenia than noncandidate genes. Biol Psychiatry 2017;82:702–708. 10.1016/j.biopsych.2017.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guloksuz S, van Os J.. The slow death of the concept of schizophrenia and the painful birth of the psychosis spectrum. Psychol Med 2017;1–16. [DOI] [PubMed] [Google Scholar]
- 9. McCarthy-Jones S. The concept of schizophrenia is coming to an end – here’s why. The Conversation 2017;2017. [Google Scholar]
- 10. Westphal J. The Mind–Body Problem. Cambridge, MA: MIT Press; 2016. [Google Scholar]
- 11. Cuthbert BN. Research domain criteria: toward future psychiatric nosologies. Dialogues Clin Neurosci 2015;17(1):89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016;3:160018 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Longo DL, Drazen JM.. More on data sharing. N Engl J Med 2016;374(19):1896–7. [DOI] [PubMed] [Google Scholar]
- 14. Greene CS, Garmire LX, Gilbert JA, et al. Celebrating parasites. Nat Genet 2017;49(4):483–4. 10.1038/ng.3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jagodnik KM, Koplev S, Jenkins SL, et al. Developing a framework for digital objects in the Big Data to Knowledge (BD2K) commons: report from the Commons Framework Pilots workshop. J Biomed Inform 2017;71:49–57. 10.1016/j.jbi.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001;69(3):89–95. 10.1067/mcp.2001.113989 [DOI] [PubMed] [Google Scholar]
- 17. Weickert CS, Weickert TW, Pillai A, et al. Biomarkers in schizophrenia: a brief conceptual consideration. Dis Markers 2013;35(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis J, Maes M, Andreazza A, et al. Towards a classification of biomarkers of neuropsychiatric disease: from encompass to compass. Mol Psychiatry 2015;20(2):152–3. 10.1038/mp.2014.139 [DOI] [PubMed] [Google Scholar]
- 19. Arranz MJ, Munro J, Sham P, et al. Meta-analysis of studies on genetic variation in 5-HT2A receptors and clozapine response. Schizophr Res 1998;32(2):93–9. 10.1016/S0920-9964(98)00032-2 [DOI] [PubMed] [Google Scholar]
- 20. Kaddurah-Daouk R, McEvoy J, Baillie R, et al. Impaired plasmalogens in patients with schizophrenia. Psychiatry Res 2012;198(3):347–52. 10.1016/j.psychres.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 21. Stevenson JM, Reilly JL, Harris MS, et al. Antipsychotic pharmacogenomics in first episode psychosis: a role for glutamate genes. Transl Psychiatry 2016;6(2):e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yao JK, Condray R, Dougherty GG Jr., et al. Associations between purine metabolites and clinical symptoms in schizophrenia. PLoS One 2012;7(8):e42165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Czerwensky F, Leucht S, Steimer W.. MC4R rs489693: a clinical risk factor for second generation antipsychotic-related weight gain?. Int J Neuropsychopharmacol 2013;16(9):2103. (9). [DOI] [PubMed] [Google Scholar]
- 24. McEvoy J, Baillie RA, Zhu H, et al. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PLoS One 2013;8(7):e68717.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh D, Poisson LM.. “Omics” data and levels of evidence for biomarker discovery. Genomics 2009;93(1):13–6. [DOI] [PubMed] [Google Scholar]
- 26. McDermott JE, Wang J, Mitchell H, et al. Challenges in biomarker discovery: combining expert insights with statistical analysis of complex omics data. Expert Opin Med Diagn 2013;7(1):37–51. 10.1517/17530059.2012.718329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MRI, U.S.D.C.f.F. Structural MRI imaging. 2017. http://fmri.ucsd.edu/Howto/3T/structure.html (10 September 2017, date last accessed).
- 28. Jia P, Han G, Zhao J, et al. SZGR 2.0: a one-stop shop of schizophrenia candidate genes. Nucleic Acids Res 2017;45(D1):D915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Essen DC, Ugurbil K, Auerbach E, et al. The Human Connectome Project: a data acquisition perspective. Neuroimage 2012;62(4):2222–31. 10.1016/j.neuroimage.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pedrosa E, Shah A, Tenore C, et al. β-catenin promoter ChIP-chip reveals potential schizophrenia and bipolar disorder gene network. J Neurogenet 2010;24(4):182–93. [DOI] [PubMed] [Google Scholar]
- 31. Allen Institute https://www.alleninstitute.org (9September 2017, date last accessed).
- 32. Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron 2010;68(2):182–6. 10.1016/j.neuron.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akbarian S, Liu C, Knowles JA, et al. The PsychENCODE project. Nat Neurosci 2015;18(12):1707–12. 10.1038/nn.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohno-Machado L, Sansone SA, Alter G, et al. Finding useful data across multiple biomedical data repositories using DataMed. Nat Genet 2017;49(6):816–9. 10.1038/ng.3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar R, Domrachev M, Lash AE.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30(1):207–10. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Omberg L, Ellrott K, Yuan Y, et al. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nat Genet 2013;45(10):1121–6. 10.1038/ng.2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krumholz HM, Waldstreicher J.. The Yale open data access (YODA) project–a mechanism for data sharing. N Engl J Med 2016;375(5):403–5. 10.1056/NEJMp1607342 [DOI] [PubMed] [Google Scholar]
- 38. Karczewski KJ, Weisburd B, Thomas B, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 2017;45(D1):D840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lonsdale J, Thomas J, Salvatore M, et al. T., The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45(6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 2007;39(10):1181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lappalainen I, Almeida-King J, Kumanduri V, et al. The European Genome-phenome archive of human data consented for biomedical research. Nat Genet 2015;47(7):692–5. 10.1038/ng.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frazier JA, Hodge SM, Breeze JL, et al. Diagnostic and sex effects on limbic volumes in early-onset bipolar disorder and schizophrenia. Schizophr Bull 2007;34(1):37–46. 10.1093/schbul/sbm120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jia P, Zhao Z.. Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum Genet 2014;133(2):125–38. 10.1007/s00439-013-1377-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 2008;40(7):827–34. 10.1038/ng.171 [DOI] [PubMed] [Google Scholar]
- 45. Ambite JL, Tallis M, Alpert K, et al. SchizConnect: virtual data integration in neuroimaging. Data Integr Life Sci 2015;9162:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Poldrack RA, Gorgolewski KJ.. OpenfMRI: open sharing of task fMRI data. Neuroimage 2017;144(Pt B):259–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kennedy DN, Haselgrove C, Riehl J, et al. The NITRC image repository. Neuroimage 2016;124(Pt B):1069–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Repovš G, Barch DM.. Working memory related brain network connectivity in individuals with schizophrenia and their siblings. Front Hum Neurosci 2012;6:137 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dolgin E. This is your brain online: the Functional Connectomes Project. Nat Med 2010;16(4):351.. 10.1038/nm0410-351b [DOI] [PubMed] [Google Scholar]
- 50. Calhoun VD, Sui J, Kiehl K, et al. Exploring the psychosis functional connectome: aberrant intrinsic networks in schizophrenia and bipolar disorder. Front Psychiatry 2011;2:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hanlon FM, Houck JM, Pyeatt CJ, et al. Bilateral hippocampal dysfunction in schizophrenia. Neuroimage 2011;58(4):1158–68. 10.1016/j.neuroimage.2011.06.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mayer AR, Ruhl D, Merideth F, et al. Functional imaging of the hemodynamic sensory gating response in schizophrenia. Hum Brain Mapp 2013;34(9):2302–12. 10.1002/hbm.22065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson A, Cohen MS.. Decreased small-world functional network connectivity and clustering across resting state networks in schizophrenia: an fMRI classification tutorial. Front Hum Neurosci 2013;7:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu Y, Yao YG, Luo XJ.. SZDB: a database for schizophrenia genetic research. Schizophr Bull 2017;43(2):459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jia P, Sun J, Guo AY, et al. SZGR: a comprehensive schizophrenia gene resource. Mol Psychiatry 2010;15(5):453–62. 10.1038/mp.2009.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Torkamani A, Dean B, Schork NJ, et al. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res 2010;20(4):403–12. 10.1101/gr.101956.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pidsley R, Viana J, Hannon E, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol 2014;15(10):483.. 10.1186/s13059-014-0483-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. O'Dushlaine C, Kenny E, Heron E, et al. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry 2011;16(3):286–92. [DOI] [PubMed] [Google Scholar]
- 59. Jia P, Wang L, Fanous AH, et al. Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS Comput Biol 2012;8(7):e1002587.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guo AY, Sun Jia JP, et al. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst Biol 2010;4:10 10.1186/1752-0509-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Prabakaran S, Swatton JE, Ryan MM, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry 2004;9(7):684–97. 643. [DOI] [PubMed] [Google Scholar]
- 62. Ng B, White CC, Klein HU, et al. An xQTL map integrates the genetic architecture of the human brain's transcriptome and epigenome. Nat Neurosci 2017;20(10):1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sullivan PF, Lin D, Tzeng JY, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry 2008;13(6):570–84. 10.1038/mp.2008.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. International Schizophrenia C, Purcell SM, Wray NR, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460(7256):748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009;460(7256):753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature 2009;460(7256):744–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015;18(2):199–209. 10.1038/nn.3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Q, Yu H, Zhao Z, et al. EW_dmGWAS: edge-weighted dense module search for genome-wide association studies and gene expression profiles. Bioinformatics 2015;31(15):2591–4. 10.1093/bioinformatics/btv150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gilman SR, Iossifov I, Levy D, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011;70(5):898–907. 10.1016/j.neuron.2011.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jia P, Zheng S, Long J, et al. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics 2011;27(1):95–102. 10.1093/bioinformatics/btq615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Richards AL, Jones L, Moskvina V, et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry 2012;17(2):193–201. 10.1038/mp.2011.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Duan F, Duitama J, Al Seesi S, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med 2014;211(11):2231–48. 10.1084/jem.20141308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest AR, Kawaji H,, et al. A promoter-level mammalian expression atlas. Nature 2014;507(7493):462–70. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Ripke S, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013;45(9):984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511(7510):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pardiñas AF, Holmans P, Pocklington AJ, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and maintained by background selection. bioRxiv 2016. doi: 10.1101/068593. [Google Scholar]
- 77. Kim Y, Xia K, Tao R, et al. A meta-analysis of gene expression quantitative trait loci in brain. Transl Psychiatry 2014;4(10):e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Colantuoni C, Lipska BK, Ye T, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 2011;478(7370):519–23. 10.1038/nature10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Aguet F, Brown AA, Castel S, et al. Local genetic effects on gene expression across 44 human tissues. bioRxiv 2016. doi: 10.1101/074450. [Google Scholar]
- 80. Zhao Z, Xu J, Chen J, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry 2015;20(5):563–72. 10.1038/mp.2014.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tao R, Davis K, Li NC, et al. GAD1 alternative transcripts and DNA methylation in human prefrontal cortex and hippocampus in brain development, schizophrenia. Mol Psychiatry 2017, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tao R, Cousijn H, Jaffe AE, et al. Expression of ZNF804A in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder. a novel transcript fetally regulated by the psychosis risk variant rs1344706 . JAMA Psychiatry 2014;71(10):1112–20. 10.1001/jamapsychiatry.2014.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kunii Y, Hyde TM, Ye T, et al. Revisiting DARPP-32 in postmortem human brain: changes in schizophrenia and bipolar disorder and genetic associations with t-DARPP-32 expression. Mol Psychiatry 2014;19(2):192–9. 10.1038/mp.2012.174 [DOI] [PubMed] [Google Scholar]
- 84. Bigos KL, Mattay VS, Callicott JH, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry 2010;67(9):939–45. 10.1001/archgenpsychiatry.2010.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li M, Jaffe AE, Straub RE, et al. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med 2016;22(6):649–56. [DOI] [PubMed] [Google Scholar]
- 86. Sekar A, Bialas AR, de Rivera H, et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016;530(7589):177–83. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fromer M, Roussos P, Sieberts SK, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 2016;19(11):1442–53. 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Takata A, Ionita-Laza I, Gogos JA, et al. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron 2016;89(5):940–7. 10.1016/j.neuron.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016;48(5):481–7. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 90. Weickert CS, Fung SJ, Catts VS, et al. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Mol Psychiatry 2013;18(11):1185–92. 10.1038/mp.2012.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Juraeva D, Haenisch B, Zapatka M, et al. Integrated pathway-based approach identifies association between genomic regions at CTCF and CACNB2 and schizophrenia. PLoS Genet 2014;10(6):e1004345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Guan L, Wang Q, Wang L, et al. Common variants on 17q25 and gene-gene interactions conferring risk of schizophrenia in Han Chinese population and regulating gene expressions in human brain. Mol Psychiatry 2016;21(9):1244–50. 10.1038/mp.2015.204 [DOI] [PubMed] [Google Scholar]
- 93. Duan J, Shi J, Fiorentino A, et al. A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. Am J Hum Genet 2014;95(6):744–53. 10.1016/j.ajhg.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cross-Disorder Group of the Psychiatric Genomics, C., Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013;381(9875):1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Thompson PM, Stein JL, Medland SE, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 2014;8(2):153–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Novak NM, Stein JL, Medland SE, et al. EnigmaVis: online interactive visualization of genome-wide association studies of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) consortium. Twin Res Hum Genet 2012;15(3):414–8. 10.1017/thg.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van Erp TG, Hibar DP, Rasmussen JM, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 2016;21(4):585.. 10.1038/mp.2015.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jaffe AE, Gao Y, Deep-Soboslay A, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2015;19(1):40–7. 10.1038/nn.4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Montano C, Taub MA, Jaffe A, et al. Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study. JAMA Psychiatry 2016;73(5):506–14. 10.1001/jamapsychiatry.2016.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lu AT, Hannon E, Levine ME, et al. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat Commun 2017;8:15353 10.1038/ncomms15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hannon E, Spiers H, Viana J, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci 2016;19(1):48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gagliano SA, Ptak C, Mak DYF, et al. Allele-Skewed DNA modification in the brain: relevance to a schizophrenia GWAS. Am J Hum Genet 2016;98(5):956–62. 10.1016/j.ajhg.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Fullard JF, Giambartolomei C, Hauberg ME, et al. Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum Mol Genet 2017;26(10):1942–51. 10.1093/hmg/ddx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li Z, Chen Yu JH, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet 2017;49:1576–83. 10.1038/ng.3973 [DOI] [PubMed] [Google Scholar]
- 105. Hall D, Huerta MF, McAuliffe MJ, et al. Sharing heterogeneous data: the national database for autism research. Neuroinformatics 2012;10(4):331–9. 10.1007/s12021-012-9151-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Fischbach GD, Lord C.. The Simons simplex collection: a resource for identification of autism genetic risk factors. Neuron 2010;68(2):192–5. 10.1016/j.neuron.2010.10.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.