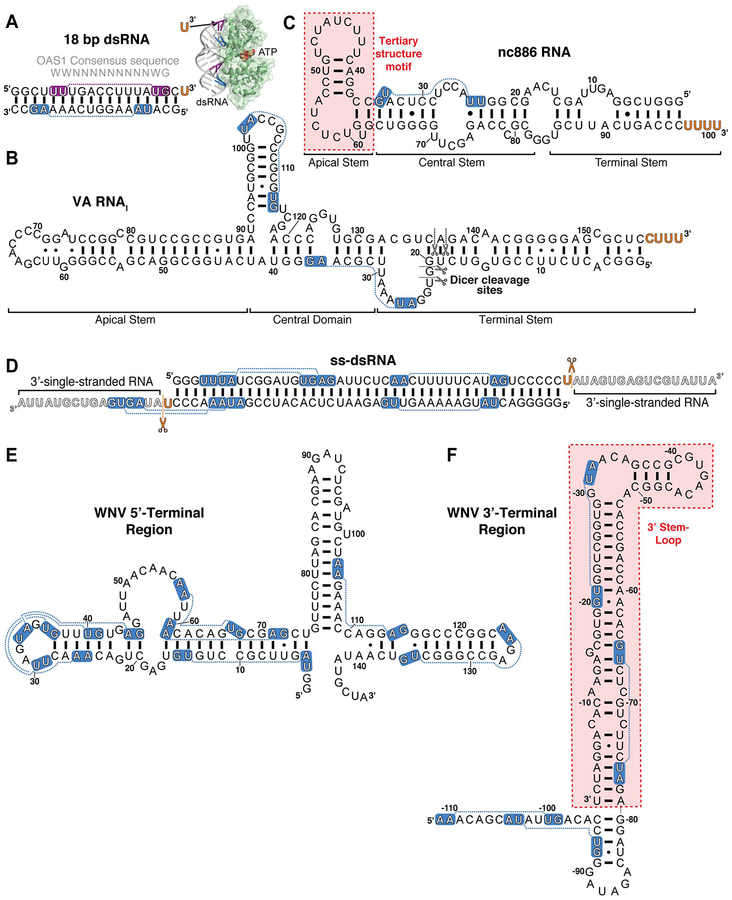
Figure 3. Double-stranded and structured RNA activators of OAS.
Select RNAs discussed in the main text: A. 18 bp dsRNA used in the OAS1-dsRNA crystal structure and identification of the 3’-ssPy motif, B. Adenovirus (Ad2) VA RNAI with sites of Dicer cleavage indicated (black scissors), C. cellular nc886 RNA with location of OAS1 activating structural motif indicated (red shading), D. Example ss-dsRNA (43 bp) with sites of RNase cleavage to generate a minimal 3’-ssPy motif indicated (orange scissors), E. WNV 5’-terminal region, and F. WNV 3’-terminal region with OAS1 activating stem-loop highlighted (red shading). For all RNAs 3’-ssPy motifs are indicated by the orange text and the OAS1 consensus activation sequence WWN9WG (shown above the 18 bp dsRNA in panel A) by white text on blue background. The purple background for the 18 bp dsRNA top strand denotes the consensus sequence directly contacted by OAS1 in the OAS1-dsRNA crystal structure (inset in Panel A).