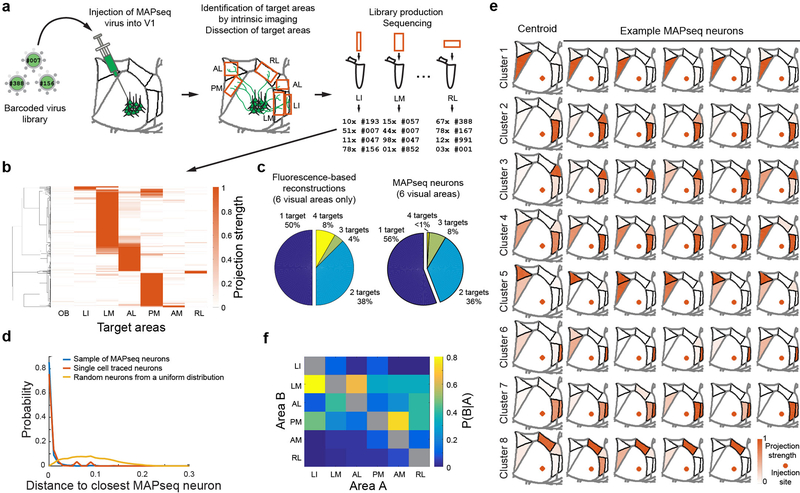
Figure 2: MAPseq projection mapping reveals a diversity of projection motifs.
(a) Overview of the MAPseq procedure. Six target areas were chosen for analysis: LI, LM, AL, PM, AM and RL. (b) Projection strength in the six target areas, as well as the olfactory bulb (OB) as a negative control, of 553 MAPseq-mapped neurons. Projection strengths per neuron are defined as the number of barcode copies per area, normalized to the efficiency of sequencing library generation and to the neuron’s maximum projection strength (n=4 mice). (c) Number of projection targets of V1 neurons when considering the six target areas only, based on the fluorescence-based axonal reconstructions (left) or the MAPseq data (right). (d) Distribution of cosine distances obtained by a bootstrapping procedure (1000 repeats) between MAPseq neurons (blue), fluorescence-based single neuron reconstructions and MAPseq neurons (orange), or random neurons (with projection strengths sampled from a uniform distribution) and MAPseq neurons (yellow). The distance distributions obtained from MAPseq neurons and fluorescence-based single-neuron reconstructions are statistically indistinguishable (Kolmogorov-Smirnov one-sided two sample test; p=0.94; α=0.05), whereas the distributions obtained from both MAPseq neurons or fluorescence-based reconstructed neurons are statistically different form the distribution obtained using random neurons (Kolmogorov-Smirnov two sample test; p<10−3; α=0.05). (e) Centroids and example cells for eight clusters obtained by k-means clustering of all MAPseq cells using a cosine distance metric. Target areas are coloured to indicate the projection strength of the plotted neuron. Projections strengths are normalize as in (b). (f) The probability of projecting to one area (Area A) given that the same neuron is projecting to another area (Area B) based on the MAPseq dataset.