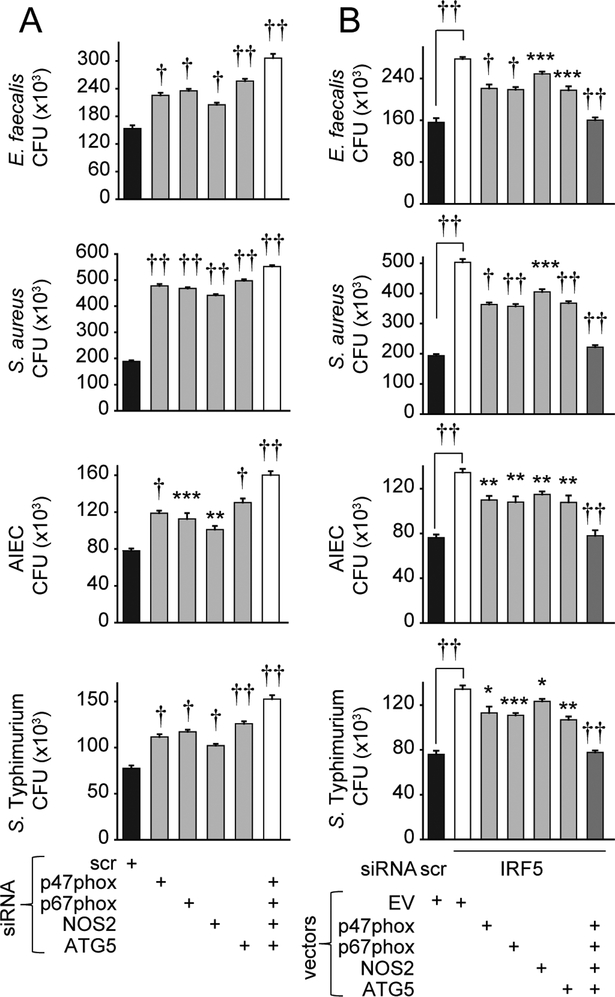
Figure 5. ROS, RNS and autophagy pathways cooperate for optimal IRF5-dependent bacterial clearance.
(A) MDMs (n=4 donors) were transfected with scrambled or the indicated siRNA, polarized into M1 macrophages, then treated with 0.1μg/ml LPS for 48h. (B) MDMs (n=8) were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA±p47phox-, p67phox-, NOS2-, or ATG5-expressing vectors alone or in combination, or empty vector (EV), then treated with 0.1μg/ml LPS for 48h. (A-B) Cells were cultured with E. faecalis, AIEC, S. aureus and S. Typhimurium. CFU+SEM. Significance is compared in ‘A’ to scrambled siRNA-transfected cells, or in ‘B’ to IRF5 siRNA, EV-transfected cells or as indicated. Scr, scrambled. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.