FIGURE 2.
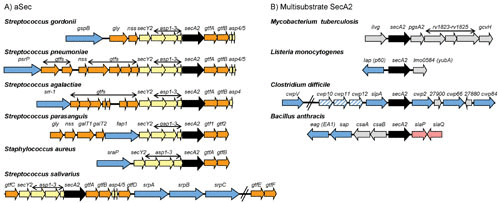
Genomic regions encoding aSec and multisubstrate SecA2 proteins. (A) aSec loci. Shown are representative aSec loci in Gram-positive bacteria. The secA2 gene is shown in black and the other genes encoding core components of the aSec translocase (SecY2 and Asps) are colored yellow. Genes encoding glycosyltransferases (Gtf) and proteins involved in carbohydrate modifications are shown in orange. Genes encoding exported SecA2 substrates are shown in blue. In Streptococcus parasanguis, the Asp orthologues are called Gap1 to Gap3. In Streptococcus salivarius, the gtfEF genes are located distal to the secA2 locus but are required for the first step of O-GlcNAcylation of the substrate (89) and thus may be functionally analogous to the gtfAB pairs found in other aSec loci. (B) Multisubstrate SecA2 loci. Shown are representative multisubstrate secA2 genes and neighboring genomic regions in Gram-positive bacteria. The secA2 gene is shown in black, and genes encoding SecA2 substrates are shown in blue. Candidate genes for additional SecA2 substrates are shown with blue stripes. Substrates encoded elsewhere in the genome are not shown. Additional proteins with roles in SecA2-dependent export are encoded by genes shown in pink. Genes encoding proteins with no known connection to export are shown in gray.
