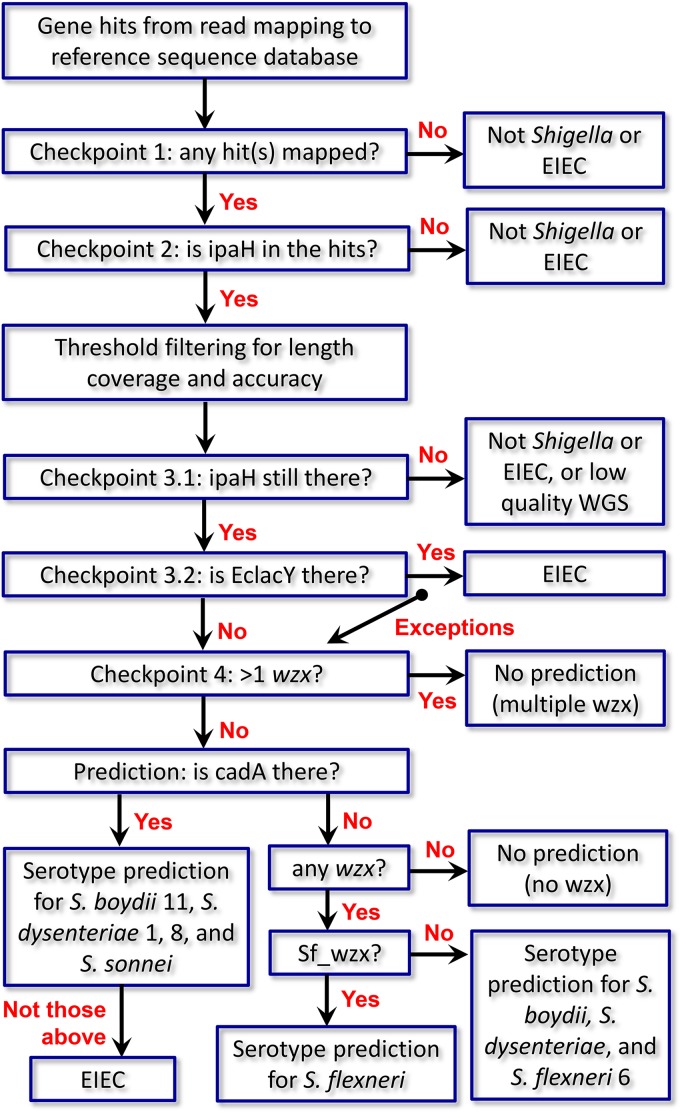
FIG 2.
Schematic illustration of a decision tree for Shigella differentiation before serotype prediction employed in ShigaTyper. ShigaTyper was designed to differentiate and exclude non-Shigella or contaminated isolates before predicting serotype for Shigella isolates. Distantly related non-Shigella/EIEC species (such as Listeria) usually have no read mapped to any of the genes in the reference sequence database and fail at checkpoint 1. Enterobacterial species (such as Salmonella) may have one or more hits but not ipaH_C and fail at checkpoint 2. Checkpoint 3 excludes EIEC based on the presence of full-length EclacY gene, with the exception of S. boydii 9 and 15. Last, if there are more than one wzx genes present in the WGS reads, it indicates multiple serotypes and fails checkpoint 4. Details on serotype prediction are provided in Results.