Figure 2.
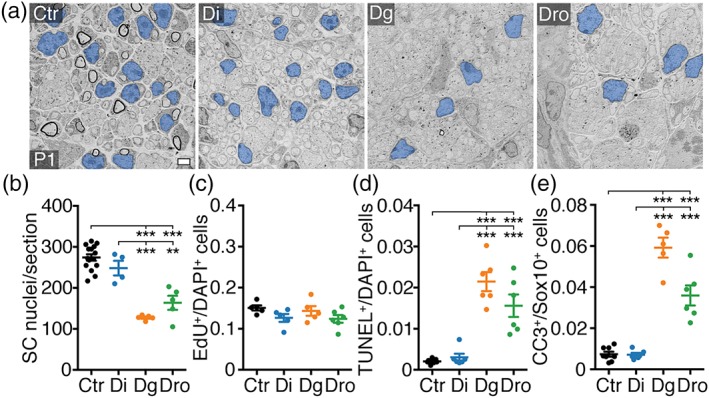
The microprocessor is required to establish correct SC numbers during radial sorting. (a) Electron micrographs of SNs of controls (Ctr), Dhh Cre ‐Dicer1 flox/flox (Di), Dhh Cre ‐Dgcr8 flox/flox (Dg), and Dhh Cre ‐Drosha flox/flox (Dro) at P1. SC nuclei are false colored in blue. (b) Number of SC nuclei per SN cross‐section in Ctr, Di, Dg, and Dro at P1. Number (n) of animals per condition: Ctr (n = 15), Di (n = 4), Dg (n = 5), Dro (n = 5). Ctr are also represented in Supporting Information, Figure S2a. (c) Number of EdU‐positive per DAPI‐positive cells in SNs of Ctr, Di, Dg, and Dro at P1. Number (n) of animals per condition: Ctr (n = 5), Di (n = 5), Dg (n = 5), and Dro (n = 6). (d) Number of TUNEL‐positive per DAPI‐positive cells in SNs of Ctr, Di, Dg, and Dro at P1. Number (n) of animals per condition: Ctr (n = 7), Di (n = 6), Dg (n = 6), and Dro (n = 6). (e) Number of cleaved caspase 3 (CC3)‐positive per Sox10‐positive SCs in SNs of Ctr, Di, Dg, and Dro at P1. Number (n) of animals per condition: Ctr (n = 8), Di (n = 6), Dg (n = 5), and Dro (n = 6). p(Dg vs Dro) = .0004. Scale bar, 2 μm (a). Error bars: SEM. One‐way anova with Tukey's multiple comparison test *p < .05, **p < .01, ***p < .001 (b–e) [Color figure can be viewed at wileyonlinelibrary.com]
