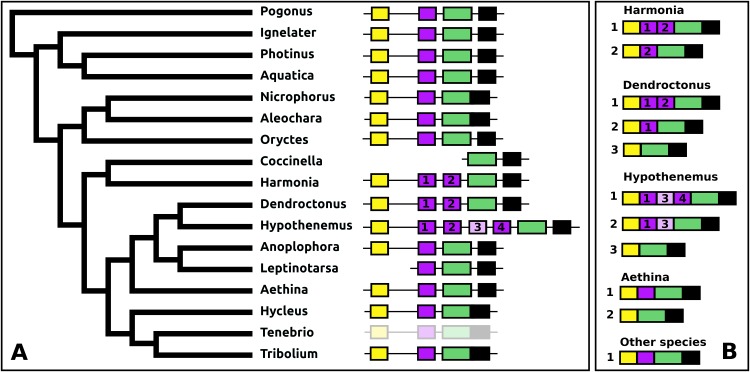
Figure 13. Structure of periviscerokinin genes.
(A) Periviscerokinin gene structures. (B) Periviscerokinin transcript structures. Horizontal lines indicate non-translated DNA and the boxes coding exons. Yellow shows the first coding exon containing the sequence coding the signal peptide and parts of the precursor, the purple coding exons contain sequences for a periviscerokinin, and the green coding exon sequences for a periviscerokinin, a tryptopyrokinin and another periviscerokinin. The final exon, black in the figure, characteristically codes for several acidic amino acid residues. In general there are relatively few RNAseq reads for this gene and when there are gaps in the genome assembly, as is the case in Leptinotarsa and Harmonia, it is not possible to reconstruct the complete gene. The structure of the Tenebrio gene is translucent to indicate that it is only inferred from those of Hycleus and Tribolium based on the very similar sequences of their periviscerokinin transcripts. To the right are the various transcripts that are produced from these genes by alternative splicing. Note that there may well be additional transcripts that could not be identified due to the scarcity of RNAseq reads for this gene.