Figure 1.
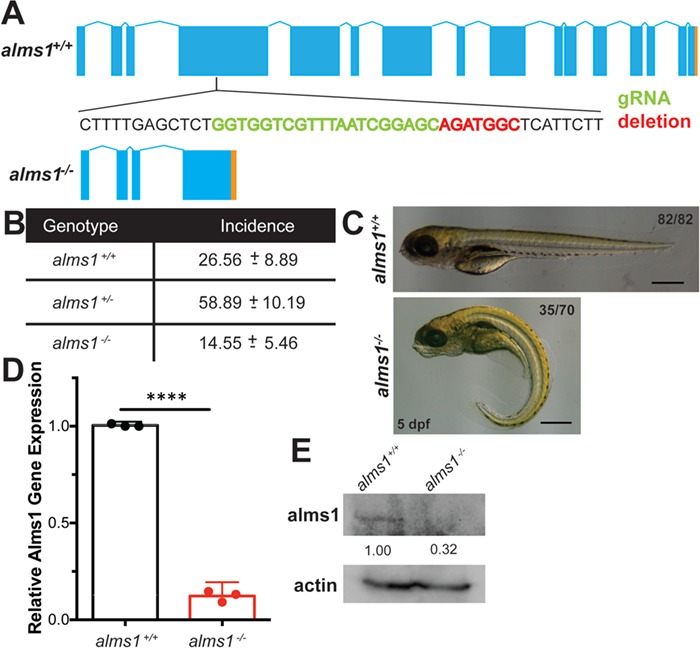
Generation of zebrafish alms1−/− line. (A) Schematic of alms1 genomic region with and without CRISPR/Cas9-induced deletion resulting in premature stop codon (orange). Sequence shown indicates region in exon 4 with sgRNA target (green) and deletion (red). (B) Ratios of identified mutants in heterozygous in-cross progeny indicating reduced rate of homozygous mutants relative to expected Mendelian ratios. (C) Representative images of alms1+/+ (n = 82) and alms1−/− (n = 70) larvae at 5 dpf. Numbers of alms1−/− larvae having ciliopathy body dysmorphogenesis shown. Scale bar, 1 mm. (D)alms1−/− larvae have reduced RNA expression levels compared to alms1+/+ at 5 dpf (n = 60–90 larvae). Statistics, Student's t-test, ****P < 0.0001. Dots, replicates; error bars, 95% confidence interval (CI). (E)alms1−/− larvae have reduced protein levels compared to alms1+/+ at 5 dpf.
