Figure 1.
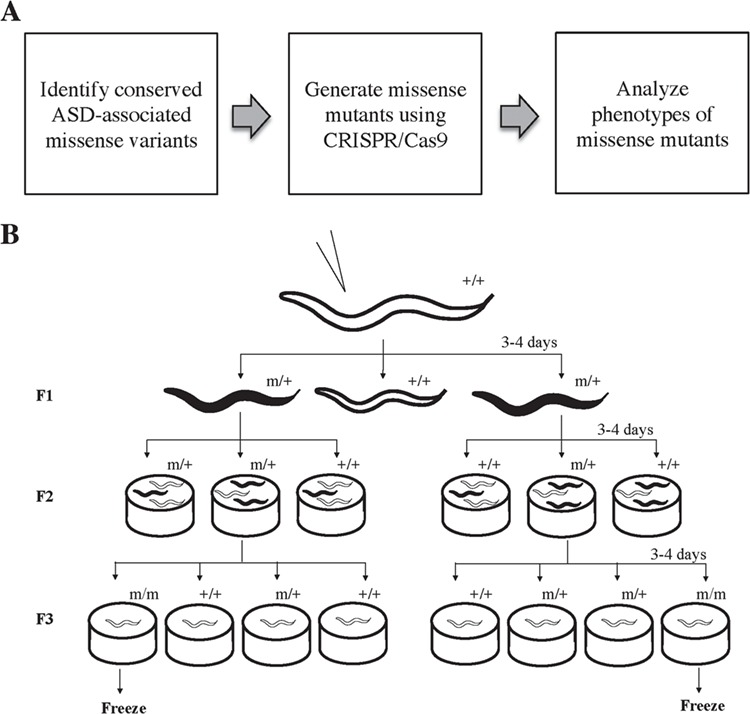
Generation of ASD-associated missense variants in C. elegans. (A) Experimental pipeline. First, we used bioinformatics to identify human ASD-associated missense variants conserved in C. elegans protein sequences. We generated the C. elegans missense mutants using CRISPR/Cas9 and homology-directed knock-in technique. We then analyzed the effects of these autism-associated alleles by comparing observable phenotypes from these alleles to both wild-type and known loss-of-function mutant controls. (B) Mutant strain screening process. Three days after injection (P0), F1 offspring expressing co-conversion marker, such as dumpy (black), were selected into individual plates. We genotyped the F2 offspring to identify the heterozygous plates. For each heterozygous plate containing the successful knock-in target missense residue (m), we randomly selected 16–20 F2 wild type-looking offspring (white) and separated them into individual plates (F3). A second round of genotyping process was then conducted to identify homozygous plates, which were cryo-preserved for future studies.
