Fig. 1.
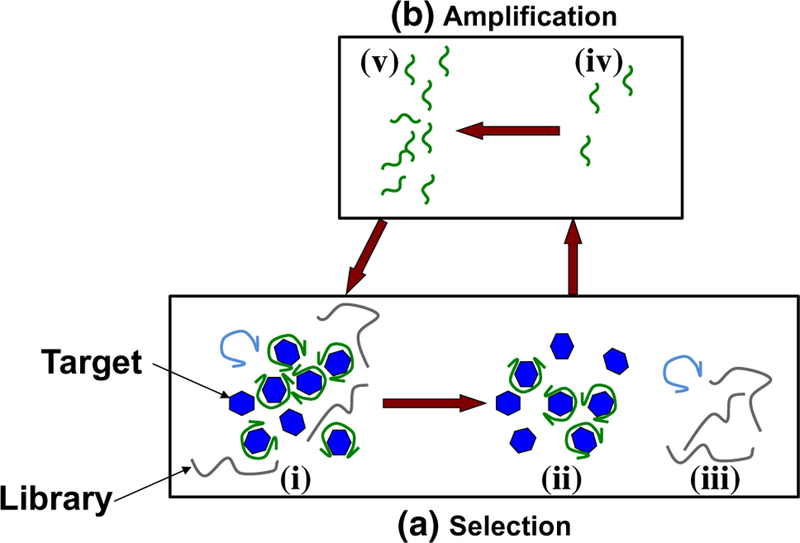
Aptamer isolation process. a A library of approximately 1014–1015 nucleic acid oligomers with randomized sequences is introduced to target molecules (i) during affinity selection. Aptamer candidates strongly bound to targets (ii) are then separated from unwanted non-binding and weakly bound nucleic acids (iii). b Aptamer candidates (iv) are then chemically amplified (v), and the process is repeated. In subsequent rounds of SELEX, these chemically amplified candidates become the new library, and the relatively weakly binding constituents are then removed during selection
