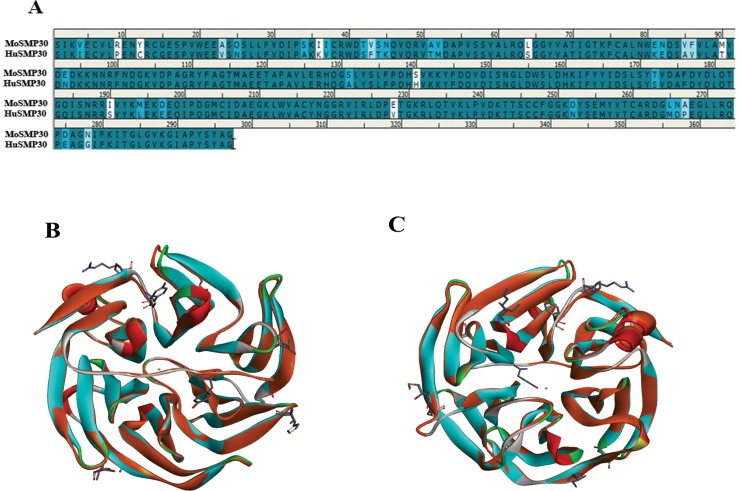
Fig 1. In silico analysis of MoSMP30 and HuSMP30.
(A) Sequence alignment of MoSMP30 and HuSMP30 protein shows 88.6% sequence identity and 95.3% sequence similarity. (B) Superimposition of MoSMP30 (Cyan) and HuSMP30 (Orange) shows minor deviation in Cα level (RMSD = 0.52Å). The dissimilar amino acid residues are presented by stick model. Cyan dot in the center represents the metal, as SMP30 is a metalloprotein (C) The top view of the superimposed crystal structure of Mouse and Human SMP30. The crystal structures were retrieved from the RCSB PDB database, and the accession ID for MoSMP30 and HuSMP30 are 4GN7 and 3G4E respectively. The analysis was performed in Discovery Studio 4.0.