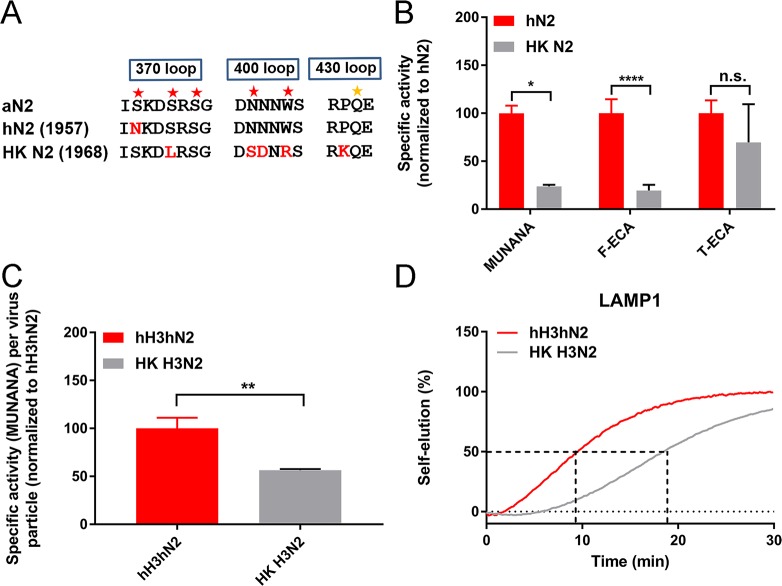
Fig 6. Enzymatic activity of HK N2 protein.
(A) Sequences of the three loops that make up the 2SBS of aN2, hN2 and HK N2 are shown. Red asterisks indicate SIA-contact residues in N2. The orange asterisk indicates an additional SIA-contact residue in the 430 loop of N9. Residues that differ from the aN2 sequence, which corresponds to the avian N2 consensus sequence, are shown in red. (B) Specific activity of hN2 and HK N2 recombinant soluble proteins as determined using the MUNANA assay or ELLA using the Fetuin-ECA (F-ECA) combination are graphed relative to hN2. (C) Relative NA activity (determined by MUNANA assay) per virus particle is graphed for hH3hN2 and HK H3N2 viruses. (B and C) Mean values of three independent experiments are shown. Standard deviations are indicated. Stars depict P values calculated using an unpaired two-tailed Student t test (*, P<0.05; **, P<0.01; ****, P<0.0001). (D) BLI analysis of virion self-elution from LAMPI-coated sensors was analysed for hH3hN2 and HK H3N2 virus particles. Prior to self-elution in the absence of OC, viruses were bound to similar levels to LAMP1 in the presence of OC. The experiment was performed three times, a representative experiment is shown.