Figure 1. Effect of dmpi8 splicing efficiency on daytime sleep requires 0.9 protein function but not dPER.
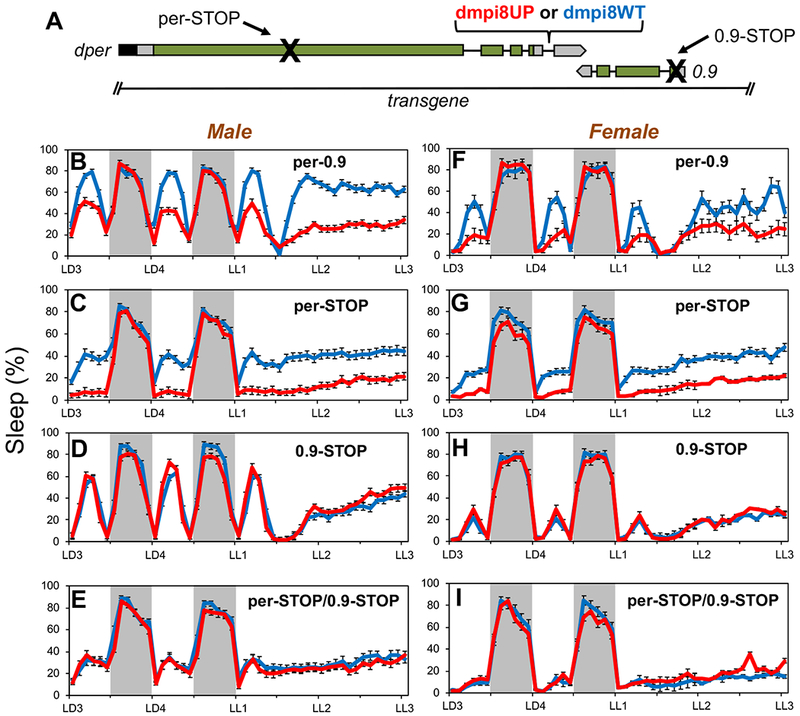
(A) Schematic of the dmpi8WT and dmpi8UP transgenes [10] modified to include a premature stop codon in the open reading frame of dper, 0.9, or both. The transgenes are based on a dper cDNA/genomic hybrid and include approximately 825bp 5’ upstream of the 0.9 transcription unit, which is transcribed in the opposite direction to dper. The 3’ untranslated regions of both dper and 0.9 overlap by 49bp. Rectangles; black (dper circadian regulatory sequence), gray (untranslated regions), green (coding region); black lines (introns). (B-G) Young adult male (B-E) or female (F-I) flies were exposed to 4 days of 12hr light/12 hr dark cycles (LD) followed by three days of continuous light (LL) at 25°C. For each genotype, graphs represent data from 96 individual flies (three independent transgenic lines × 32 individual flies/line) and data pooled to get the population average. Shown are the relative sleep levels during the last 2 days of LD followed by 2 days of LL for transgenic flies carrying the dmpi8UP (red) or dmpi8WT (blue) versions of the wildtype per-0.9 (B, F) per-STOP (C, G), 0.9-STOP (D, H) or double stop (E, I). Grey rectangle, 12 hr dark period. The following lines were used to obtain group averages; dmpi8WT, kx-f4c, m38-k, f9; dmpi8UP, m17, m32, f13; per-STOP[dmpi8WT], m53, f18, f46; per-STOP[dmpi8UP], m55, m131, m138; 0.9-STOP[dmpi8WT], m50, m62, m73; 0.9-STOP[dmpi8UP], m55, f50, f59; per/0.9-STOP[dmpi8WT], m94, m100, f38; per/0.9-STOP[dmpi8UP], m42, m81, f5. Representative examples are shown and similar results were obtained with other independent lines (see STAR Methods and KEY RESORCE TABLE for identity of lines analyzed). See also Figures S1 and S2 for additional results.
