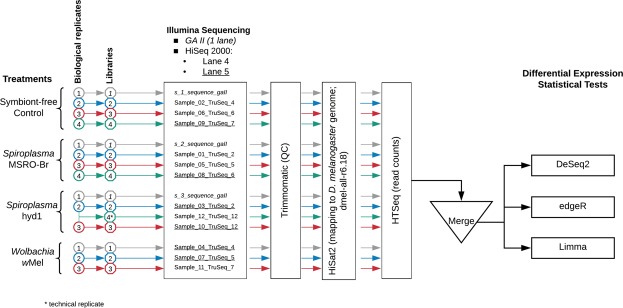
Figure 1.
Experimental design and workflow for data analysis. The biological replicates, corresponding libraries, and sequence file labels are distinguished by colours and font type. Italics = samples run on Illumina GAII. Non-italics = samples run on Illumina HiSeq 2000; non-underlined samples were pooled into one sequencing lane (4), whereas underlined samples were pooled into another lane (5). Library labelled “Sample_12_TruSeq_12” is the result of combining total RNA from Hyd1 biological replicates 2 and 3, and thus considered a technical, rather than biological, replicate. This library was excluded from the differential expression analyses.