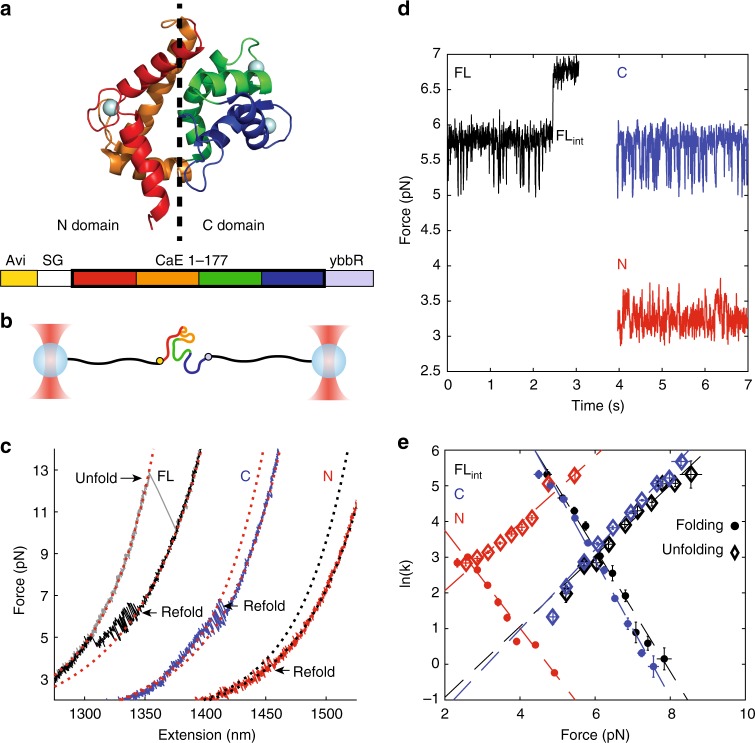
Fig. 1.
Full-length calerythrin folds through the C domain. a The NMR structure of calerythrin (PDB: 1NYA20). EF 1, 2, 3, and 4 are red, orange, green, and blue, respectively. The protein sequence contains an Avi tag at the N terminus and ybbR tag at the C-terminus. b The geometry in OT is shown, with 2 kb of DNA on each side of the protein. c Force–extension curves of the FL, N, and C domain samples, offset in extension. The WLC model of DNA+protein is shown as dotted red lines, except for N domain, which is dotted black. The protein WLCs correspond to lengths of 177, 75, and 90 amino acids for FL, C, and N. The FL protein unfolds cooperatively (gray) and refolds in two steps (black). The C (blue) and N (red) domains fold reversibly in one step; for clarity only the refolding curves are shown. d Passive of the FL, C, and N constructs (offset in time). In all cases, the lowest observed force is the unfolded state, and then transitions are observed to a folded state. For FL (black), the first transition is FLint and the highest transition is the full-folded structure. The N and C fold to their one-domain structures. e The kinetics of folding (circles) and unfolding (diamonds) for FLint match the kinetics of the C domain but are markedly different than the N domain. Error bars are standard error (SE). FLint n = 11 molecules, 116 rate measurements; C domain n = 9 molecules, 107 rate measurements; N domain n = 13 molecules, 100 rate measurements. See Supplementary Table 1 for the fits of the Bell model (dashed lines). See Supplementary Fig. 2 for further confirmation of FLint identity. Source data are provided as a Source Data file