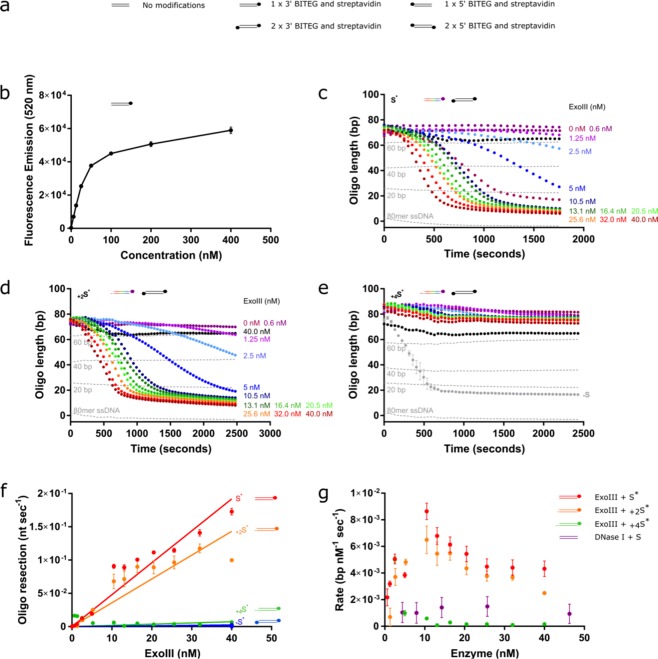
Figure 2.
Assay validates inhibitory effect of increasing lengths of a 3′ overhang on 3′-5′ nuclease, ExoIII. (a) Key to stick and ball illustrations of the substrates. The parallel horizontal lines (=) represent the dsDNA substrate, and the coloured-in circle represents the combination of both biotin and streptavidin (●). (b) Calibration curve depicting the fluorescent signal according to increasing concentrations of 80-bp oligomer substrate with one terminal BITEG modification and in the presence of 0.02 mg/ml streptavidin. (c–e) ExoIII titrations as depicted in the figures on 50 nM substrates presenting a blunt terminus (c), 2-nt (d) or 4-nt (e) 3′ overhang (multi-coloured cartoons), and negative controls (black cartoons) containing BITEG-modified 3′-ends. Results were normalised against their respective negative controls (absence of ExoIII) and converted to bp. The grey curve in (e) shows the equivalent reaction without an overhang, highlighting the loss of activity with a 4 bp 3′ overhang. (f) Rate of resection by ExoIII on the blunt (red), 2-nt (orange) and 4-nt (green) 3′ overhangs and the negative control with BITEG-treated 3′-ends. Minimal loss of activity is observed with a 2-nt overhang, in contrast to almost complete loss of activity with a 4-nt overhang. g, Comparison of ExoIII and DNase I activity on their respective substrates. Error bars represent SEM; n = 3 in all cases.