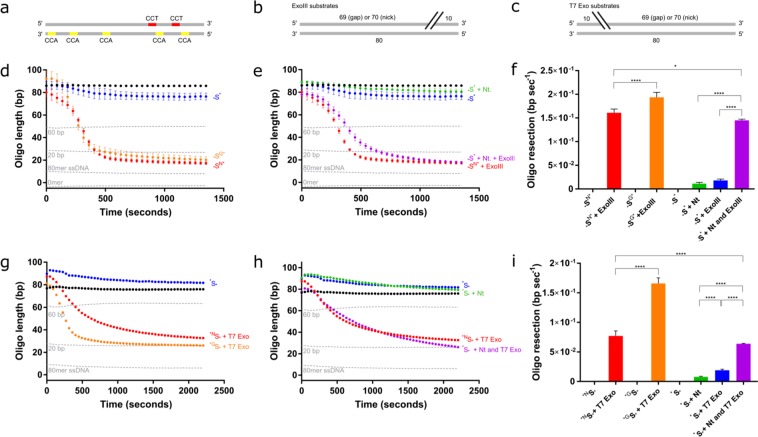
Figure 6.
ExoIII and T7 Exo preferentially resect from a nick than a gap. (a) Schematic of the dsDNA substrate indicating the preferred nicking sites (yellow) and the least favourable nicking sites (red) of Nt.CviPII. (b,c) Schematic of the dsDNA substrates presenting a nick or gap at either the 3′-end for ExoIII, or the 5′-end for T7 Exo. (d) ExoIII is active against simulated nicked substrates. 40 nM ExoIII was added to a blocked substrate (blue) and related substrates designed with either a nick (red) or a gap (orange) towards the 3′ end of one strand, showing similar activity against both modified substrates. Standard curve is represented by the grey dotted lines. (e) Quantification of the Nt.CviPII nickase enzyme activity coupled to ExoIII. Nt.CviPII was added to the blocked substrate, either with (purple) or without (green) 40 nM ExoIII. Controls from b) are shown for comparison. (f) Calculated resection rate of nicked and gapped substrates by ExoIII, extracted from the maximum gradient. Addition of the nickase significantly increases the resection rate, highlighting that the nickase activity is detected. (g) T7 Exo is active against simulated nicked substrates. 12 nM T7 Exo was added to substrates with a nick (red) or a gap (orange) towards the 5′ end of one strand. Negative control is in blue. Standard curve is represented by the grey dotted lines. (h) Quantification of the Nt.CviPII enzyme activity coupled to T7 Exo. Nt.CviPII was added to the blocked substrate, either with (purple) or without (green) 12 nM T7. Negative control is in blue. (i) Calculated resection rate of nicked and gapped substrates by T7 Exo, extracted from the maximum gradients in (g) and (h). Addition of the nickase significantly increases the resection rate, highlighting that the nickase activity is detected. Error bars represent SEM; n = 3 in all cases; *p < 0.05, ****p < 0.0001.