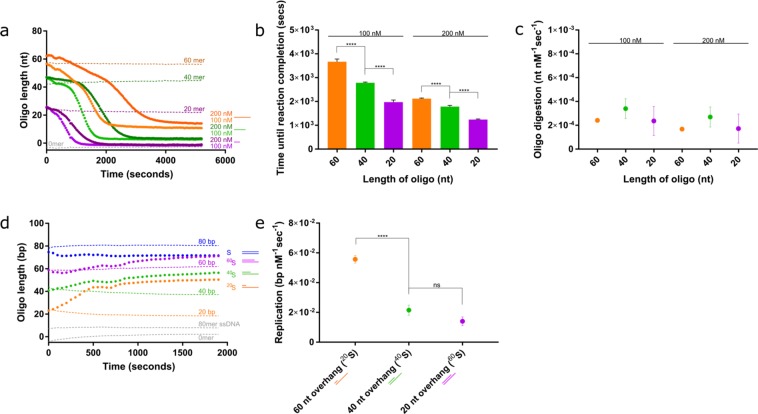
Figure 7.
Validation of the assay for alternative enzymes that digest ssDNA and generate dsDNA. (a) Mammalian Trex2 digestion of single stranded DNA quantified using the PG assay. Single-stranded 60-mer (orange), 40-mer (green) and 20-mer (purple) DNA substrates were treated with 100 and 200 nM Trex2. Robust digestion was observed in each case. (b) Time (seconds) until reaction completion based on the point at which the graphs plateau in (a). (c) Rate of digestion per nM Trex2. Trex2 degrades DNA at a similar rate irrespective of the length of the DNA substrate. (d) The polymerase activity of the Klenow fragment polymerase determined using the PG assay. Three substrates with a 20-, 40- and 60- and overhang with a total 80 bases were incubated with 1 nM Klenow fragment. (e) Calculated polymerisation rate of the Klenow fragment. A clear preference is shown for longer overhangs. Error bars represent SEM; n = 3 in all cases; ****p < 0.0001.