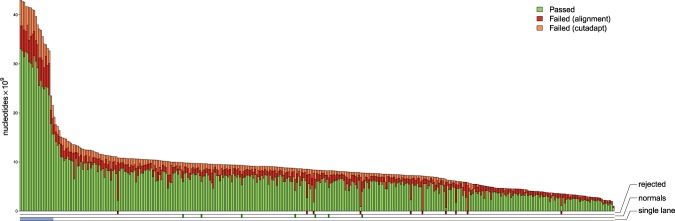
Figure 2.
Nucleotide summaries for each library. Overall, for the 335 libraries sequenced at 5 per lane, sequencing yielded an average of 82.1 × 106 reads (21.3 × 106–191.4 × 106). For the 20 libraries sequenced at 1 per lane, there was an average of 368.2 × 106 reads (215.3 × 106–425.6 × 106). The total nucleotide count (across both pairs) for each library is represented by the bar height. This is divided into nucleotides removed during preprocessing (“failed cutadapt” in orange) and nucleotides removed during alignment and read post-processing (“failed alignment” in red). The remaining nucleotides (“passed” in green) are present in the downstream analyses. Library annotations are shown below the plot. The libraries run at a higher depth at one library per lane are marked (“single lane”). The 10 libraries identified as low quality and subsequently rejected are marked (“rejected”), as are the seven libraries from normal samples (“normals”). Following pre-processing (adapter trimming, dropping of reads less than 20 bp, and quality score less than 20), a median of 79.1 × 106 (5.5 × 106–189.3 × 106) reads (multiplexed) and 366.1 × 106 (219.6 × 106–423.8 × 106) reads (single) were successfully aligned. After alignment, deduplication, and read quality filtering, a median of 65.1 × 106 (multiplexed) and 320.0 × 106 (single) reads were retained. 10 libraries which were clear outliers, showing exceptionally low alignment rates (3.1 × 106–34.3 × 106 aligned reads retained), were rejected from subsequent analyses. Overall, the non-rejected libraries gave a median alignment rate of 0.82 (0.47–0.90) yielding a median coverage of 1.8x (multiplexed) and 9.1x (single).