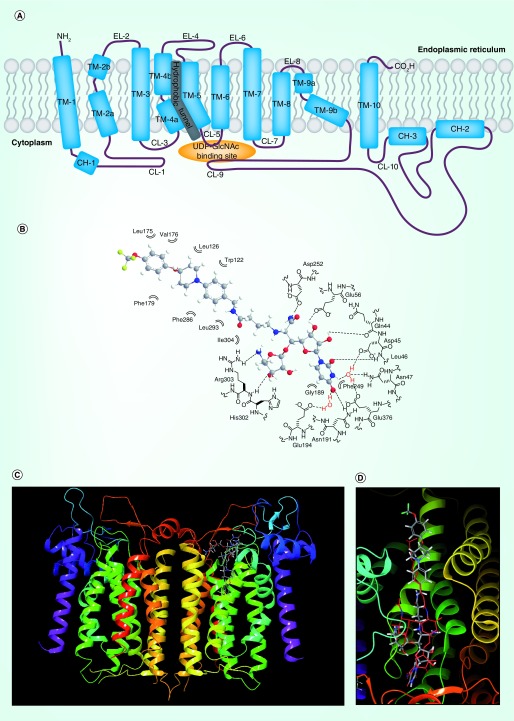
Figure 3. . Two-dimensional representation of DPAGT1 (A) and docking studies of APPB (B, C & D).
(B) Schematic representation of interactions between APPB andits DPAGT1 binding domain. (C) The superimposed structures of DPAGT1-tunicamycin and DPAGT1-APPBa. (D) Close-up view of the superimposed structures of tunicamycin and APPBa. (C & D) aAPPB (1) was dockedwith DPAGT1 in complex with tunicamycin (Protein Data Bank ID: 6BW6) using Glide on Maestro from Schrodinger. The carbons of 1 are shown in grey; the hydrogens are white; the nitrogens are blue; the fluorines are green; the oxygens are red. Tunicamycin is shown in red. The docking score was -12.57 (Schrödinger, LLC, OR, USA). These images were generated with Maestro 11.5. DPAGT1 shows a homodimerict structure.
CH: Cytoplasmic helix segment; CL: Cytoplasmic loop; EL: ER loop; TM: Transmembrane segment.