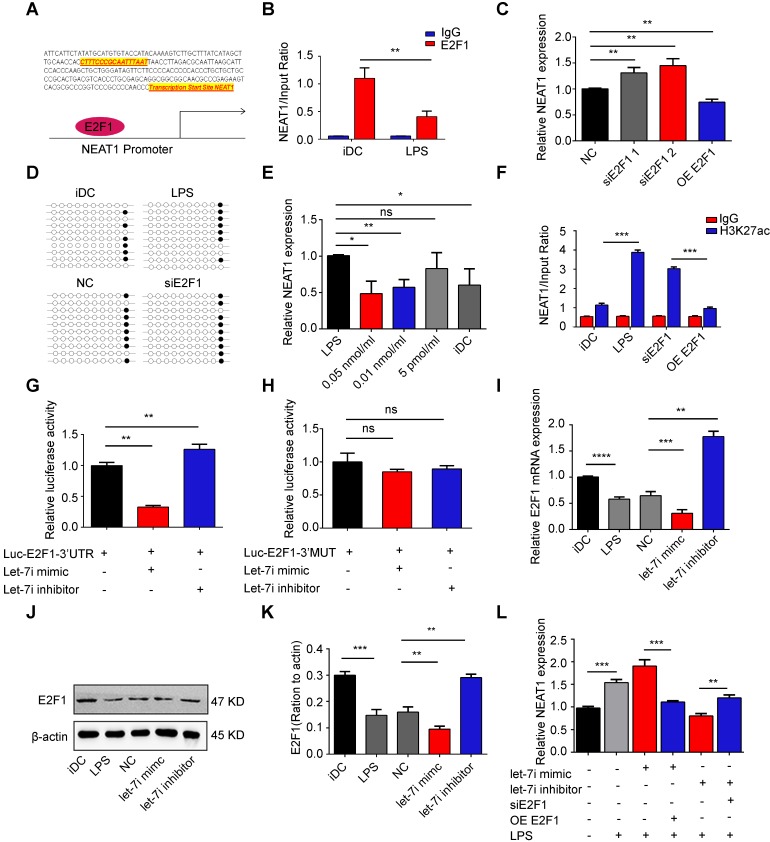
Figure 4.
NEAT1 was regulated by the let-7i/E2F1 axis. (A) The E2F1-binding elements were identified in the promoter region of NEAT1. (B) Chromatin immunoprecipitation assay was performed using E2F1 or IgG antibody in immature DCs (iDCs) and LPS-DCs. The relative expression of NEAT1 was detected by qRT-PCR. Data are expressed as mean ± SD; n = 3 biological replicates; **p<0.01, derived by Student's t-test. (C) DCs were transfected with E2F1 siRNA (siE2F1 1 or siE2F1 2), E2F1 pcDNA3.1 vector (OE E2F1), or negative control (NC) for 12 h before undergoing stimulation with LPS. The expression of NEAT1 was detected by qRT-PCR. Data are expressed as mean ± SD; n = 3 biological replicates; **p<0.01, derived by Student's t-test. (D) DCs were transfected with siE2F1 or NC for 12 h before undergoing stimulation with LPS. Methylation status of the CpG island was assessed by bisulfite sequencing in immature DCs (iDCs), LPS-DCs, NC, and siE2F1 DCs. Each row represents a single clone. (E) DCs were stimulated with Trichostatin A at different concentrations (0.05 nmol/ml, 0.01 nmol/ml, or 5 pmol/ml) for 12 h before undergoing stimulation with LPS. The expression of NEAT1 was detected by qRT-PCR. LPS-DCs were used as positive controls and iDCs were used as negative controls. Data are expressed as mean ± SD; n = 3 biological replicates; *p<0.05, **p<0.01, ns, not significant, derived using Student's t-test. (F) DCs were transfected with siE2F1 or OE E2F1 for 12 h before stimulation with LPS. Chromatin immunoprecipitation assay was performed using H3K27ac or IgG antibody in DCs. The relative expression of NEAT1 was detected by qRT-PCR. Data are expressed as mean ± SD; n = 3 biological replicates; ***p<0.001, derived using Student's t-test. (G-H) A reporter construct with potential binding site for let-7i in the 3'-UTR-wild type (WT) or 3'UTR-mutant type (MUT) region of E2F1 was generated. 293T cells were transiently transfected with miR-3076-3p mimic of miR-3076-3p inhibitor for 24 h. Luciferase activities were measured and normalized to the level of Renilla luciferase used as control (G-H). Data are expressed as mean ± SD; n = 3 biological replicates; **p<0.01 derived using Student's t-test. (I-K) DCs were transfected with a let-7i inhibitor or mimic for 12 h before stimulation with LPS. mRNA expression of E2F1 was detected by qRT-PCR (I). LRP3 protein level was detected by western blotting (J-K). Data are expressed as mean ± SD; n = 3 biological replicates; **p<0.01, ***p<0.001, derived using Student's t-test. (L) DCs were transfected with let-7i mimic, let-7i inhibitor, let-7i mimic+OE E2F1, or let-7i mimic+siE2F1 for 12 h before stimulation with LPS. The expression of NEAT1 was assessed by qRT-PCR. Data are expressed as mean ± SD; n = 3 biological replicates; **p<0.01, ***p<0.001, derived using Student's t-test.