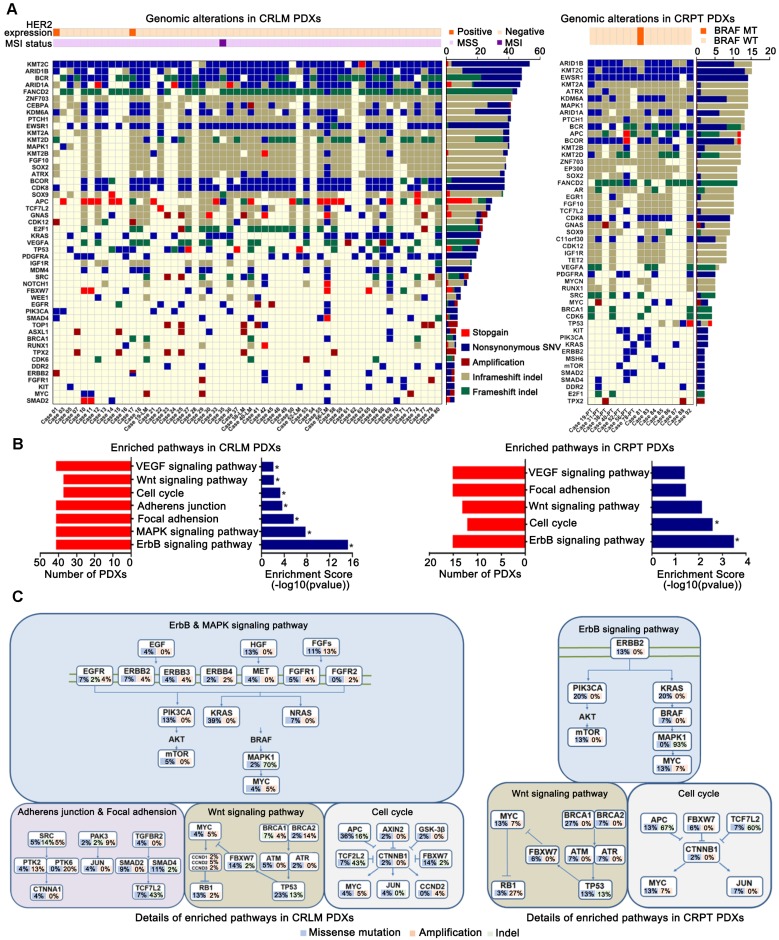
Figure 3.
Identification of variants and pathway enrichment in PDX models from patients with CRLM. (A) Genomic alterations were analyzed with different clinicopathological features in PDX models from CRLM (Left) and CRPT (Right) respectively. Red, stopgain; Dark blue, nonsynonymous SNV; Maroon, Amplification; Dark khaki, inframeshift indel; Green, frameshift indel. (B) Several relevant pathways, including ErbB, MAPK, focal adhesion, adherens junction, cell cycle, Wnt and VEGF pathways in CRLM PDXs (Left), were found to be enriched by KEGG analysis, whereas only ErbB, cell cycle signaling pathways were enriched in the CRPT PDXs (Right). * p < 0.05. (C) Details for molecular alterations involved in the KEGG analysis were showed in CRLM (Left) and CRPT (Right) PDX models respectively. Blue, missense mutation; Pale green, indel; Orange, amplification. Copy number ≥ 5 was considered as amplification.